Figure 3. Control of lncRNA production by WDR82-ZC3H4.
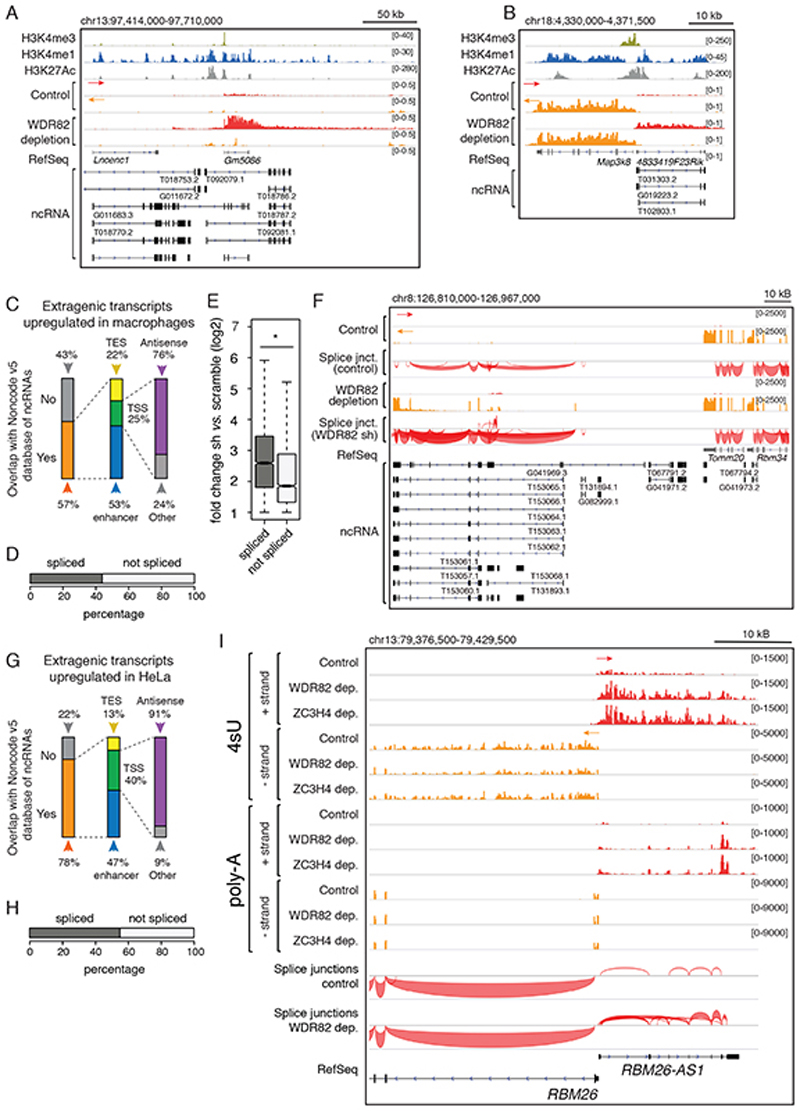
A-B) A representative candidate enhancer (A) and promoter (B) showing the overlap of WDR82-suppressed transcription with annotated lncRNAs in macrophages.
C) Overlap between WDR82-suppressed extragenic transcripts (n=2,870) in mouse macrophages and lncRNAs in the NONCODE v5 database. TSS = Transcription Start Site; TES = Transcription End Site.
D) Transcripts upregulated upon WDR82 depletion in 4sU RNA-seq data in mouse macrophages (n=2,870) were classified as spliced (n=1,292) or unspliced (n=1,578) based on polyA RNA-seq data.
E) Signal intensity of spliced and unspliced WDR82-suppressed transcripts detected in 4sU-seq data in mouse macrophages (n=4 independent experiments). The median value for each fold change is shown with a horizontal black line. Boxes show values between the first and the third quartile. The lower and upper whisker show the smallest and the highest value, respectively. Outliers are not shown. The notches correspond to ~95% confidence interval for the median. Statistical significance was assessed using the two-tailed Wilcoxon rank sum test (p-value=2.1e-92). * = p-value < 0.01.
F) PolyA-RNA-seq snapshot from mouse macrophages showing splice junctions at a representative genomic region containing lncRNAs whose expression was increased upon WDR82 depletion.
G) Overlap between transcripts upregulated in 4sU-seq datasets from HeLa cells depleted of WDR82 (n=1,509) and lncRNAs annotated in NONCODE v5.
H) Transcripts identified as upregulated in 4sU RNA-seq data in HeLa cells depleted of WDR82 (n=1,509) were classified as spliced (n = 850) or unspliced (n = 659) based on polyA RNA-seq data.
I) Representative genomic region showing a coding gene (RBM26) and the associated promoter-antisense transcription in HeLa cells depleted of WDR82 or ZC3H4. Strand-specific 4sU-seq and polyA RNA-seq data are shown.