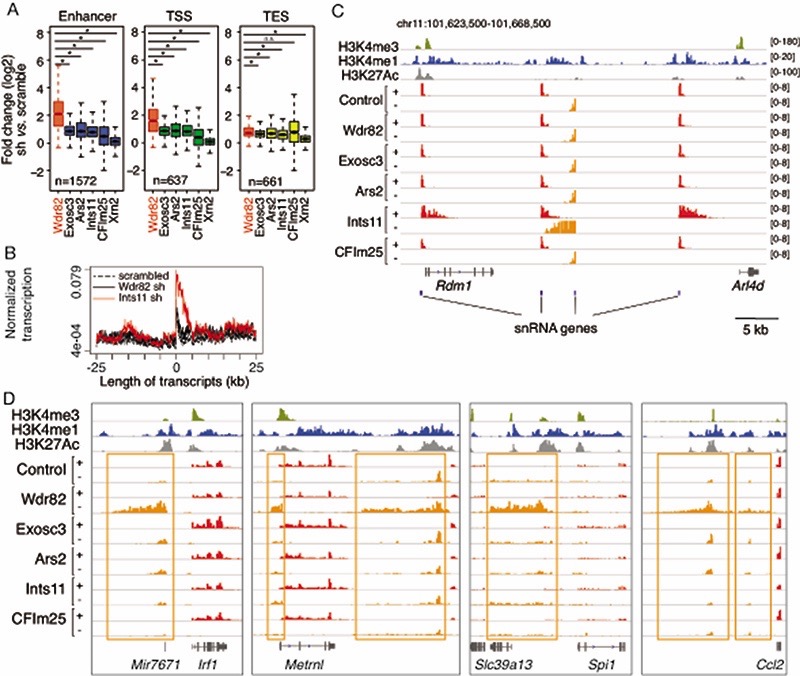
Extended Data Fig. 1. Extragenic transcription in cells depleted of WDR82 or other transcription terminators.
A) The effects of the depletion of known termination factors on extragenic transcription was measured by 4sU labeling and sequencing in mouse bone marrow-derived macrophages. We considered the n=2,870 extragenic regions whose transcription was increased in macrophages depleted of WDR82 at 45’ after LPS stimulation and measured their 4sU labeling in macrophages depleted of the indicated proteins. Each transcript was assigned to the nearest annotated enhancer, Transcription Start Site (TSS) or Transcription End Site (TES). The log2-transformed fold change (sh vs. scramble) for each depletion experiment is shown. Statistical significance was assessed using the two-tailed Wilcoxon signed rank test and a p-value ≤ 0.01 was considered significant. p-values for transcripts assigned to Enhancers: Exosc3 p-value= 2.2e-208, Ars2 p-value= 2.9e-206, Ints11 p-value= 1.4e-217, CFIm25 p-value= 4.4e-189, Xrn2 p-value= 9.2e-239. p-values for transcripts assigned to TSS: Exosc3 p-value= 4.6e-69, Ars2 p-value= 3.7e-73, Ints11 p-value= 1.6e-72, CFIm25 p-value= 7.1e-72, Xrn2 p-value= 7.8e-101. p-values for transcripts assigned to TES: Exosc3 p-value= 1.5e-14, Ars2 p-value= 5.8e-10, Ints11 p-value= 6.7e-26, CFIm25 p-value= 0.149775, Xrn2 p-value= 1.0e-81. *** = p-value <0.01. ns: not statistically significant. Inside the boxplot, the median value for each fold change is shown with a horizontal black line. Boxes show values between the first and the third quartile. The lower and upper whisker show the smallest and the highest value, respectively. Outliers are not shown. The notches correspond to ~95% confidence interval for the median.
B) Comparison of the effects of the depletion of WDR82 and INTS11 on transcription termination at snRNA genes.
C) A representative genomic region on mouse chromosome 11 containing multiple snRNA genes.
D) Snapshots of genomic regions showing the effects of the depletion of WDR82 and other termination factors on extragenic transcription.