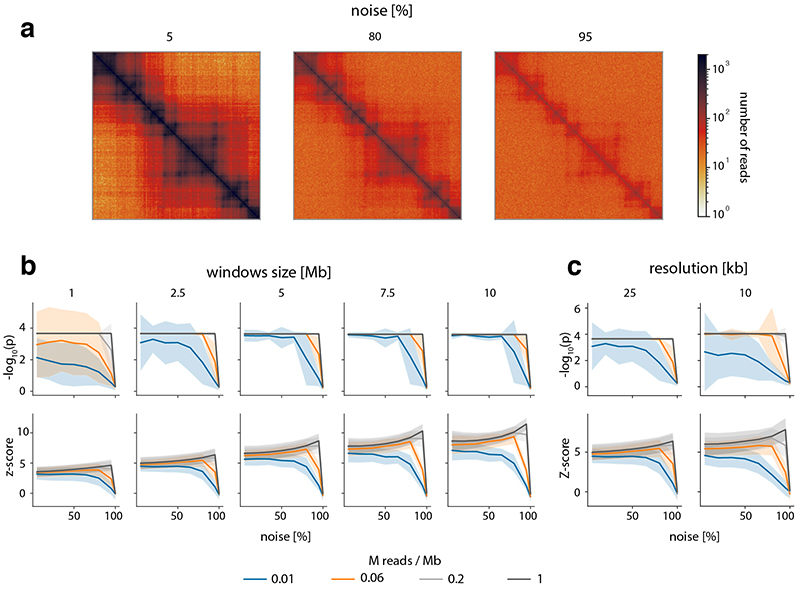
Extended Data Fig. 4. CHESS is robust to changes in noise due to random ligations and sequencing depth in real Hi-C data.
a, Examples of 5 Mb matrices used in this analysis including a 5, 80 and 95 % of added noise (random ligations between pairs of loci). We tested to what extent CHESS is able to identify two matrices as being identical, after noise and sequencing depth were adjusted independently in them. Matrices are based on chromosome 19 data from Bonev et al. 201712. a, examples of the data with different amounts of noise. b, empirically determined p-values and z-scores of CHESS runs with different window sizes, noise levels and simulated sequencing depths (details in Online Methods). Step size and matrix resolution were both 25 kb. Lines for 2 x 105 and 1 x 106 overlap for runs with window sizes > 1 Mb. c, As in panel a, but comparing CHESS runs with 2.5 Mb window size on matrices binned at 25 kb and 10 kb. b, and c, solid lines indicate the mean, shaded areas the standard deviation over 1976, 2066, 2156, 2246, 2300 matrix pairs for window sizes 10 Mb, 7.5 Mb, 5 Mb, 2.5 Mb, 1 Mb, respectively.