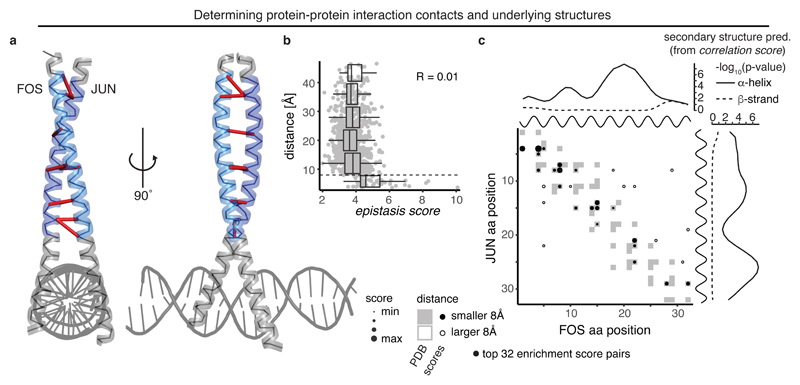
Fig. 4. Deep mutagenesis identifies protein-interaction contacts.
a, Crystal structure of the leucine zipper domains of FOS and JUN with a DNA strand (PDB entry 1fos). The mutated regions (32 amino acids each) are highlighted in light blue (FOS) and dark blue (JUN)11. Top 10 enrichment score pairs are shown with red dashes, note that two interactions between position 8 in FOS and positions 7 and 8 in JUN, as well as three interactions between positions 14 and 15 in FOS and positions 14 and 15 in JUN are hard to distinguish.
b, Distance of position pairs as a function of enrichment scores (n = 1,024). Boxplots are spaced in intervals of 8 Å; boxes cover 1st to 3rd quartile of the data, with middle bar indicating median, whiskers extend at maximum to 1.5-times the inter quartile range away from the box. Dashed horizontal line indicates 8 Å threshold. Pearson correlation coefficient is indicated.
c, FOS-JUN trans interaction score map for top 32 position pairs with highest enrichment scores, compared to contact map of known interaction structure (1fos, underlying in grey). Note that protein-protein interaction maps are not symmetric. Shown on top and to the right of the contact map are the known alpha helices (black) as well as the secondary structure propensities derived from correlation scores of FOS and JUN (one-sided permutation test, see also Supplementary Fig. 4a,b).