Fig. 6. Deep learning improves contact prediction and structural models from deep mutagenesis data.
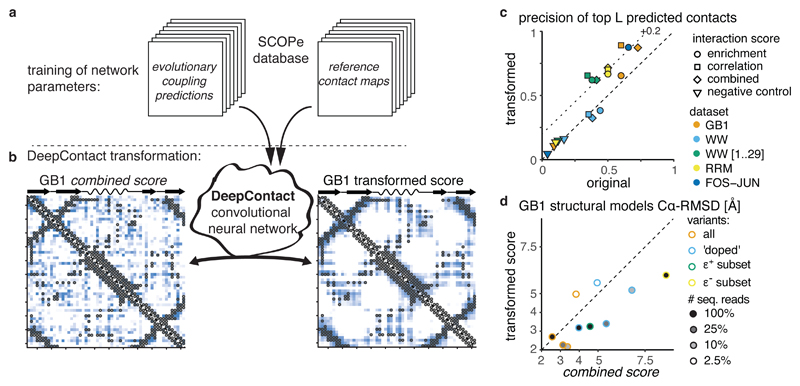
a, DeepContact convolutional neural network transforms DMS-derived interaction score maps based on learned structural patterns44. The basic DeepContact architecture used here takes as the only input the DMS-derived interaction score map and transforms it based on structural patterns previously learned on an orthogonal and independent training set (in which it compared evolutionary coupling-derived contact predictions with contacts in known structures of representative protein families in the SCOPe database).
b, GB1 domain combined score interaction map before (left panel) and after (right panel) transformation with DeepContact convolutional neural network. Heat maps show scores (low -white, high - blue). Grey open circles show contacts (distance < 8 Å) in reference structure.
c, Precision of top L predicted contacts before and after DeepContact transformation. Negative control is average over three random permutations of combined score matrices (in case of FOS-JUN dataset enrichment score matrices).
d, Comparison of accuracy 〈Cα − RMSD〉 of top 5% GB1 structural models (n = 25 each) with restraints derived either from combined scores or from DeepContact-transformed combined scores for different (down-sampled) GB1 DMS datasets.