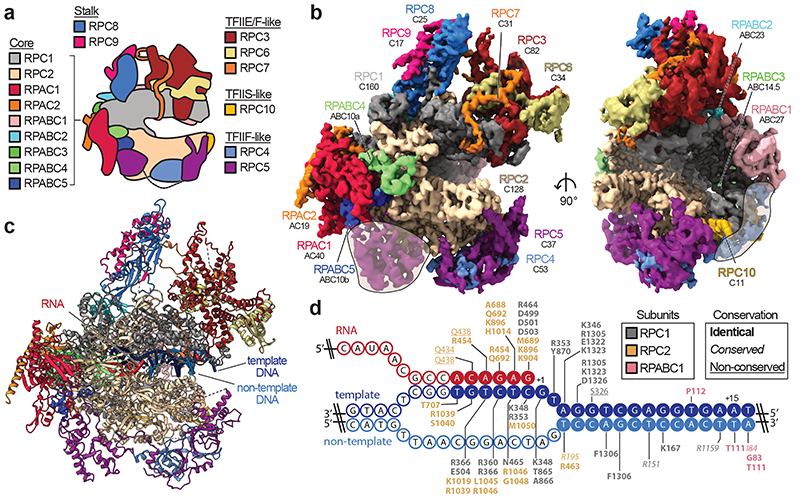
Fig. 1. Cryo-EM structure of human RNA polymerase III.
a, Schematic of human Pol III and colour code of its subunits grouped by subcomplexes or homology to Pol II factors. b, Cryo-EM map of apo human Pol III shown from the side (left) and front (right) with human subunits coloured as in a and additionally labelled with their yeast counterparts below. Shadings mark superimposed features of a cryo-EM map derived from a pooled dataset of apo and elongating Pol III. c, Structural model of elongating human Pol III with bound nucleic acids. d, Schematic of the DNA/RNA transcription scaffold. Filled and unfilled circles represent modelled and non-modelled nucleotides, respectively, labelled with contacting residues within 4 Å distance. Boxes on the top-right show the subunit colour-code and used typography of the residues to mark their conservation between human and yeast Pol III.