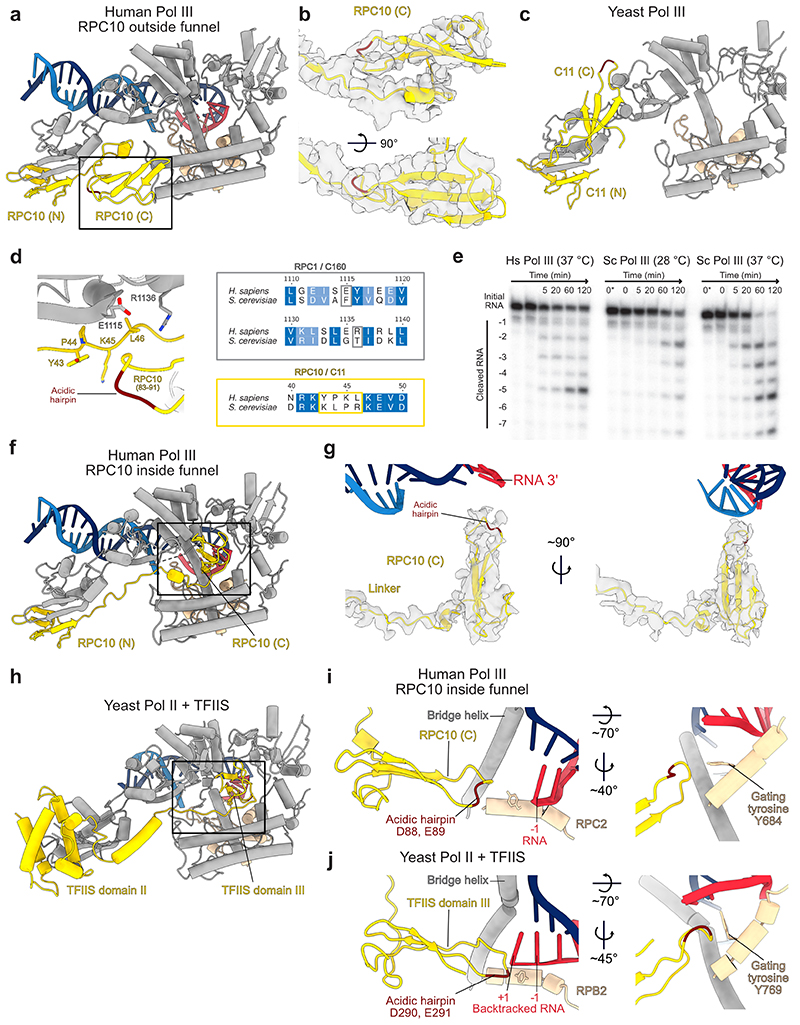
Extended Data Fig. 6. Structures of human RPC10 in different conformations.
Shown are cartoon representations of human RPC10 or its homologues in yeast, of nucleic acids and of the polymerase regions that form the pore and funnel domains and the jaw domain. Acidic hairpins of RPC10, C11 and TFIIS are coloured dark red. a, RPC10 C-ribbon (C) adopting the ‘outside funnel’ conformation in the elongating Pol III structure. b, Cryo-EM density (map E) is depicted as transparent grey surface and shows the fit of the C-ribbon of RPC10 in the ‘outside funnel’ conformation. c, C11 (the yeast homolog to RPC10) shows a different conformation of its C-ribbon domain, whereas the position of the N-ribbon (N) is similar in both species. Shown is the yeast apo Pol III structure (PDB 5fja) because the C-ribbon is not visible other conformational states. d, The RPC10 C-ribbon in the ‘outside funnel’ conformation folds back and bind its linker and the RPC1 jaw. RPC10 from residues 83 to 91 are shown as ribbon, and interacting residues within 4 Å distance are shown as sticks (left). Sequence alignments of H. sapiens RPC1 and S. cerevisiae C160 and of H. sapiens RPC10 and S. cerevisiae C11 covering the respective regions (right) shows that the contacting residues are not conserved. e, RNA cleavage assay of the 32P-labelled RNA annealed to the same transcription scaffold as used for structure determination of elongating Pol III. Shown is a time course from 0 to 120 minutes. Lane 0* contains RNA in the absence of polymerase. Hs – H. sapiens; Sc – S. cerevisiae. Uncropped image is shown in the Source Data. f, RPC10 (C) adopting the ‘inside funnel’ conformation (EC-3 Pol III) upon straightening of the linker. g, Cryo-EM density (map D) is depicted as transparent grey surface and shows the fit of the C-ribbon of RPC10 in the ‘inside funnel’ conformation. h, Structure of yeast TFIIS bound to Pol II in its ‘reactivation intermediate’ conformation (PDB 3PO3). i, j, Comparison between the structures of i, human Pol III RPC10 in the ‘inside funnel’ conformation and j, yeast Pol II bound to TFIIS (PDB 3PO3). Close-up views of RPC10 and TFIIS reaching into the active are shown on the right. RPC10 doesn’t reach as far into the polymerase active site as TFIIS. The acidic hairpin is inflected in RPC10 and is extended in TFIIS. Gating tyrosines of human Pol III RPC2 (Y684) and yeast Pol II RPB2 (Y769) are shown as sticks and adopt different conformations.