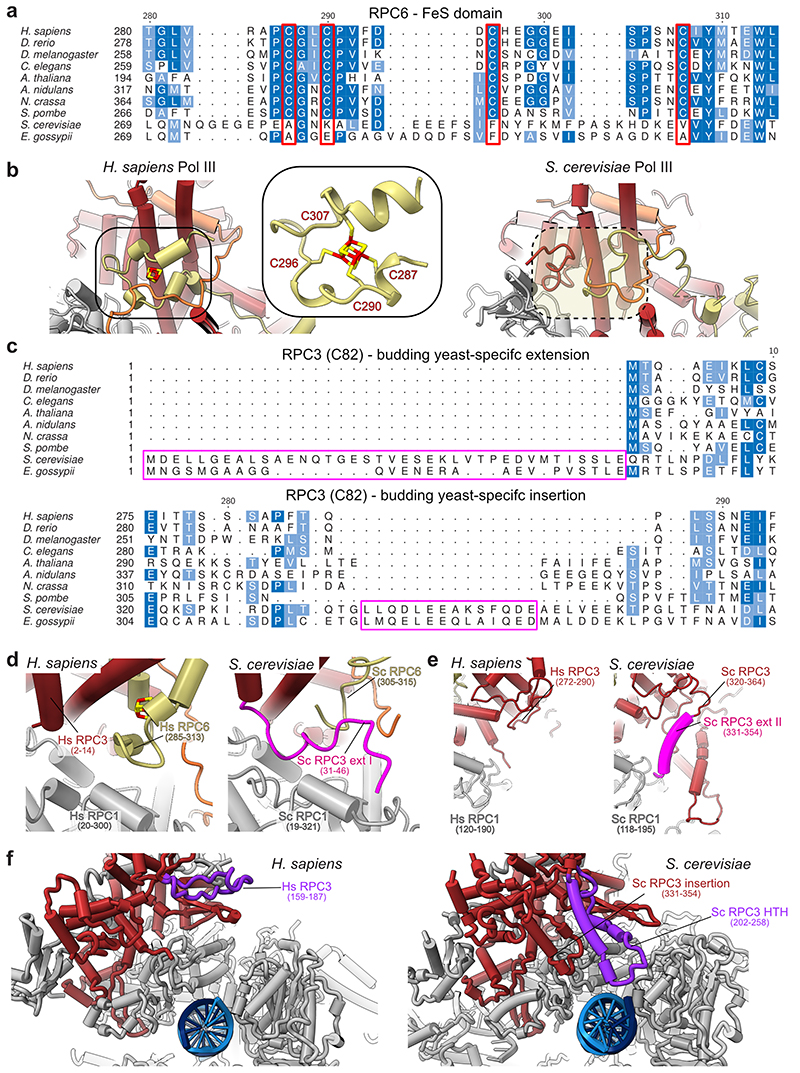
Extended Data Fig. 8. Structure, function and evolution the RPC6 FeS domain and RPC3 extensions.
a, Multiple sequence alignment (MSA) of the RPC6 C-terminal FeS domain from model organisms. MSA was generated via T-Coffee74 implemented into the EMBL-EBI bioinformatic tools web service75. The four cysteine residues required for coordination of the FeS cluster are highlighted in red. b, Comparison of the RPC3-RPC6-RPC7 heterotrimer from H. sapiens (left, elongating Pol III) and S. cerevisiae (right, PDB 6eu3). The cubane FeS cluster is coloured in red and yellow. The position of the FeS cluster is framed, and a close-up view shows the four cysteines coordinating the FeS cluster. The equivalent position in S. cerevisiae Pol III structure, which lacks the FeS cluster, is marked with a dashed oval. c, MSA of the RPC3 (C82 in S. cerevisiae) N-terminus and of region 275-294 (H. sapiens) generated with T-Coffee. The MSA of the RPC3 N-terminus was manually adjusted, based on the available structural information (see panel d), so that the N-terminus of budding yeasts becomes apparent as an N-terminal extension instead of an insertion. The budding yeast specific N-terminal extension and central insertion (331-354 in S. cerevisiae) are highlighted with pink frames. d-f, Close-up comparisons between H. sapiens (elongating Pol III) and S. cerevisiae (PDB 6eu3 in d, e and 6f41 in f) Pol III structural elements that contribute to heterotrimer integrity and anchoring to the RPC1 (C160 in S. cerevisiae) core subunit. d, Budding yeast-specific N-terminal extension is positioned similarly as the FeS cluster in H. sapiens Pol III. Notably, only 16 residues (31-46) of the extension are visible, whereas the remaining amino acids are flexible. e, The central insertion (331-354) binds the polymerase core, whereas this contact point is missing in H. sapiens. f, S. cerevisiae Pol III contains a helix-turn-helix (HTH) insertion (202-258) that binds the downstream DNA in the S. cerevisiae PIC (PDB 6f41). The HTH insertion further binds and stabilises the central insertion (331-354) and, thereby, likely contributes to the anchoring of the heterotrimer to the polymerase core. Both elements are absent in H. sapiens although the counterpart region (159-187 in H. sapiens) has a similar shape (albeit without α-helical elements) that points into a different direction. Hs – H. sapiens; Sc – S. cerevisiae. Sc RPC1, Sc RPC3, Sc RPC6 relate to Sc C160, Sc C82, Sc C34, respectively, according to the commonly used nomenclature of S. cerevisiae Pol III subunits.