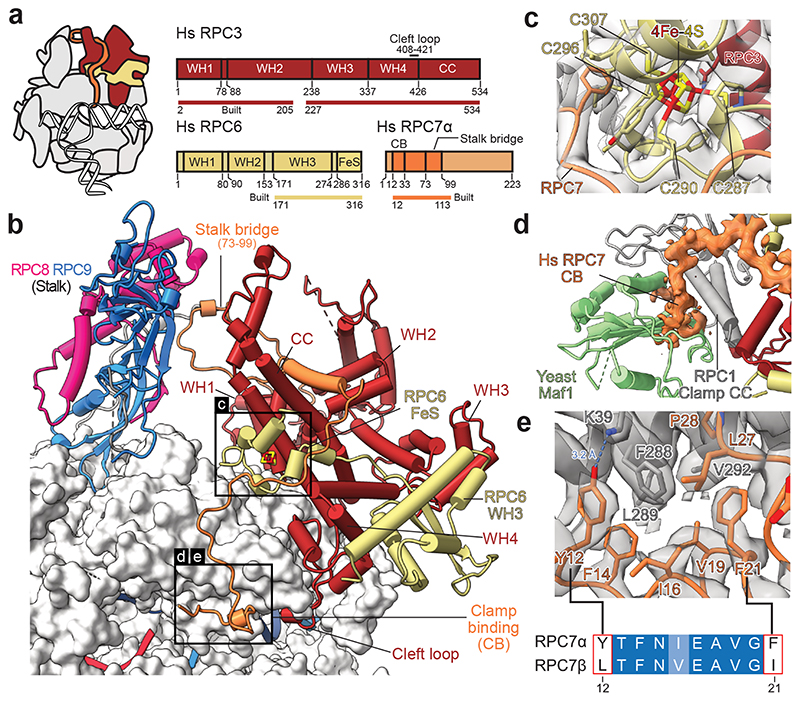
Fig. 4. Architecture of human RPC3-RPC6-RPC7 heterotrimer.
a, Schematic domain organisation of subunits RPC3, RPC6, and RPC7 in the elongating human Pol III complex. Coloured bars indicate modelled regions and are labelled as ‘built’. WH – winged helix, CC – coiled-coil, FeS – Iron-sulphur (4Fe-4S) binding, CB – clamp binding. b, Heterotrimer (RPC3-RPC6-RPC7) and stalk (RPC8-RPC9) are shown as cartoons and bind the Pol III core shown as surface. Boxed regions highlight features that are not visible in yeast Cryo-EM structures. c, Structure of the RPC6 FeS domain binding the cubane 4Fe-4S cluster. Residues within 6 Å of the FeS cluster are shown as sticks, and the four coordinating cysteines are labelled. Cryo-EM density is shown as a transparent surface. d, Superimposition of the apo human Pol III structure with the yeast Pol III-Maf1 structure (PDB 6tut). Only Maf1 is shown and is coloured light green. Cryo-EM density of RPC7 is shown as an orange surface. e, Interface between the RPC1 clamp coiled-coils (CC, grey) and RPC7 (orange). RPC1 F288 is surrounded by aromatic residues of RPC7, of which Y12 and F21 are exclusive to the tumour-associated RPC7α isoform as shown by the sequence alignment between RPC7α (UniProt: O15318) and RPC7β (UniProt: Q9BT43) placed below. Corresponding Cryo-EM density is shown as a grey transparent surface. The blue dashed line indicates a putative hydrogen bond between Y12 (RPC7) and K39 (RPC1).