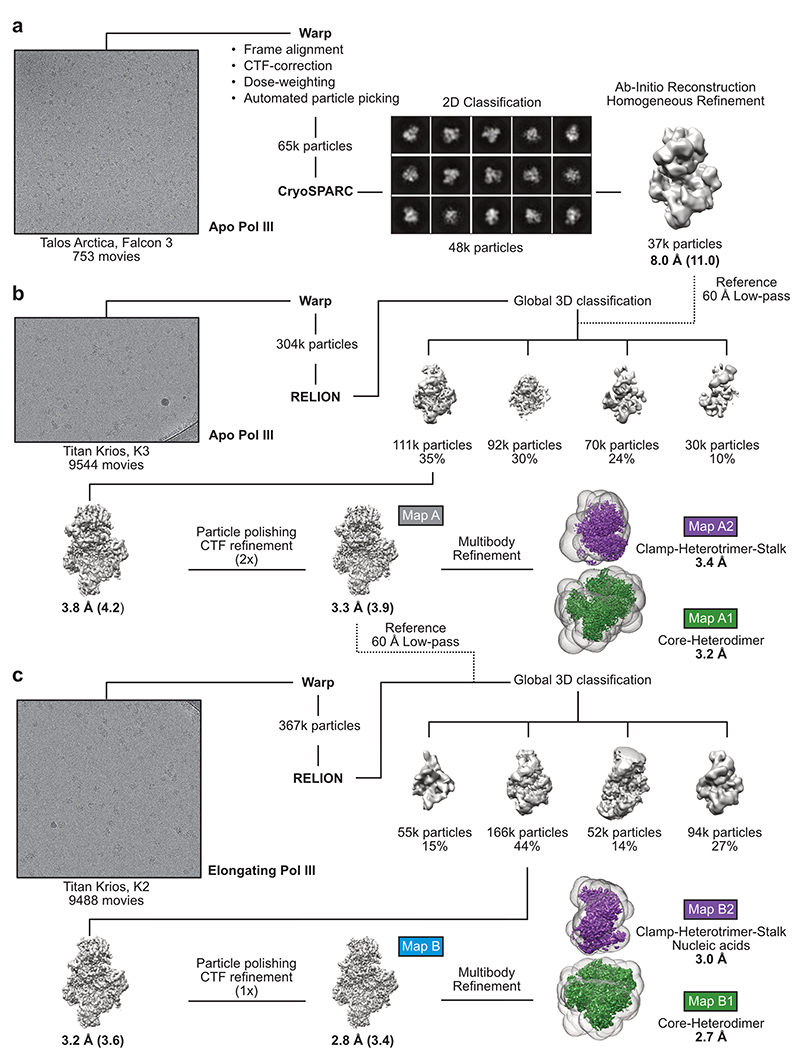
Extended Data Fig. 2. Global Cryo-EM data processing strategy of apo and elongating human Pol III.
a, Talos Arctica screening dataset, from which a low-resolution 3D reconstruction of apo human Pol III at 8 Å was obtained. b, Processing pipeline of the Titan Krios data set of apo human Pol III. For global 3D classification, the low-resolution Apo Pol III map (filtered to 60 Å) was used as a reference model. c, Processing pipeline of the Titan Krios data set of elongating human Pol III. For global 3D classification, the high-resolution Apo Pol III map (filtered to 60 Å) was used as a reference model. High-resolution cryo-EM maps were further subjected to RELION multi-body refinement67 with applied masks (transparent surfaces) covering the core and heterodimer (green maps A1, B1) and the clamp, heterotrimer and stalk domains (purple maps A2, B2). Representative micrographs of the three datasets are shown on the left. Shown are the unsharpened cryo-EM maps whereas reported resolution values correspond to automatically B-factor sharpened maps obtained via RELION post-processing. Resolution values of the unsharpened maps are added with parenthesis. Shown particle numbers were rounded down. All maps, marked with coloured boxes, were used for structural model building.