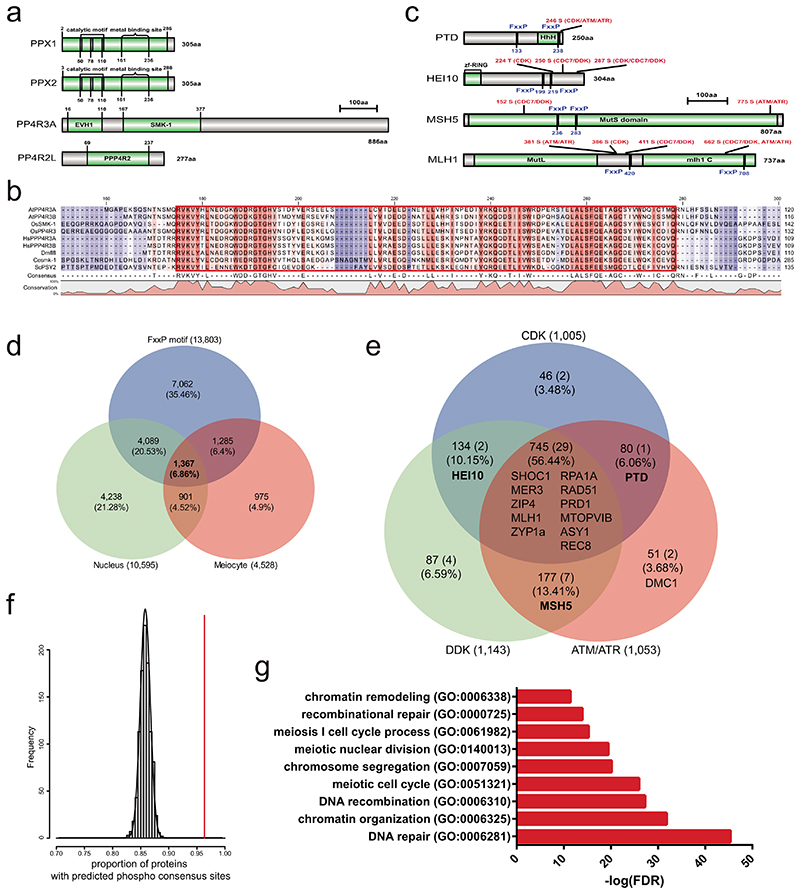
Extended Data Fig. 10. Genome-wide prediction of PP4 complex target proteins during meiosis.
a, Protein domain (green) structure of Arabidopsis PP4 subunits PPX1, PPX2, PP4R2 and PP4R3. b, Amino acid alignment of the PP4R3A homolog EVH1 domain (red box). Hash symbols (#) indicate conserved tyrosine (Y) and tryptophan (W) residues. c, As for a, but showing the positions of FxxP motifs and phosphorylation consensus sites in PTD, HEI10, MSH5 and MLH1. d, Venn diagram showing overlap of meiotically expressed, nuclear proteins with FxxP motifs. e, Venn diagram showing overlap of candidate PP4 target proteins with CDK, DDK or ATM/ATR kinase consensus motifs, predicted using GPS 5.06. The location of HCR1 Y2H interactors are indicated within the Venn diagram. f, Histogram showing a significant enrichment of proteins containing phosphorylation sites in the predicted 1,367 PP4 targets, compared to 1,000 sets of randomly chosen genes (n=1,367). The vertical red line indicates observed predicted PP4 target proteins containing phosphorylation sites, compared to the random sets (black lines). g, Gene ontology (GO) enrichment analysis of the predicted PP4 targets, using PANTHER (http://pantherdb.org/). Benjamini-Hochberg False Discovery Rate (FDR) correction was used for enrichment test.