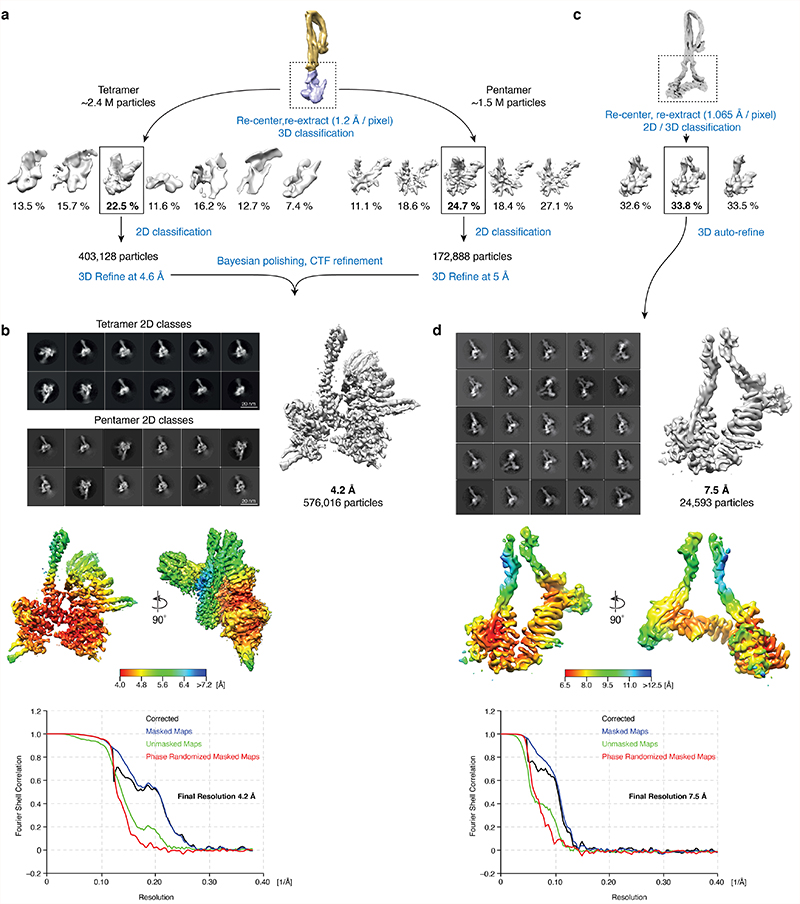
Extended Data Fig. 3. Focused image processing of the head segments of nucleotide-free apo condensin.
a, Workflow for the apo non-engaged condensin head segment. Particles of tetramer and pentamer complexes were initially processed and preliminary angles and translations were assigned as described in Extended Data Fig. 1. b, Representative 2D classes (top), 4.2 Å cryo-EM density map and its local resolution plot as calculated by RELION (middle) and FSC curves (bottom) for the non-engaged head segment. c, Workflow for the apo bridged condensin head segment. Particles of the bridged class of the pentamer complex were initially processed and preliminary angles and translations were assigned as described in Extended Data Fig. 1. d, Representative 2D classes (top), 7.5 Å cryo-EM density map and its local resolution plot as calculated by RELION (middle) and FSC curves (bottom) for the bridged head segment.