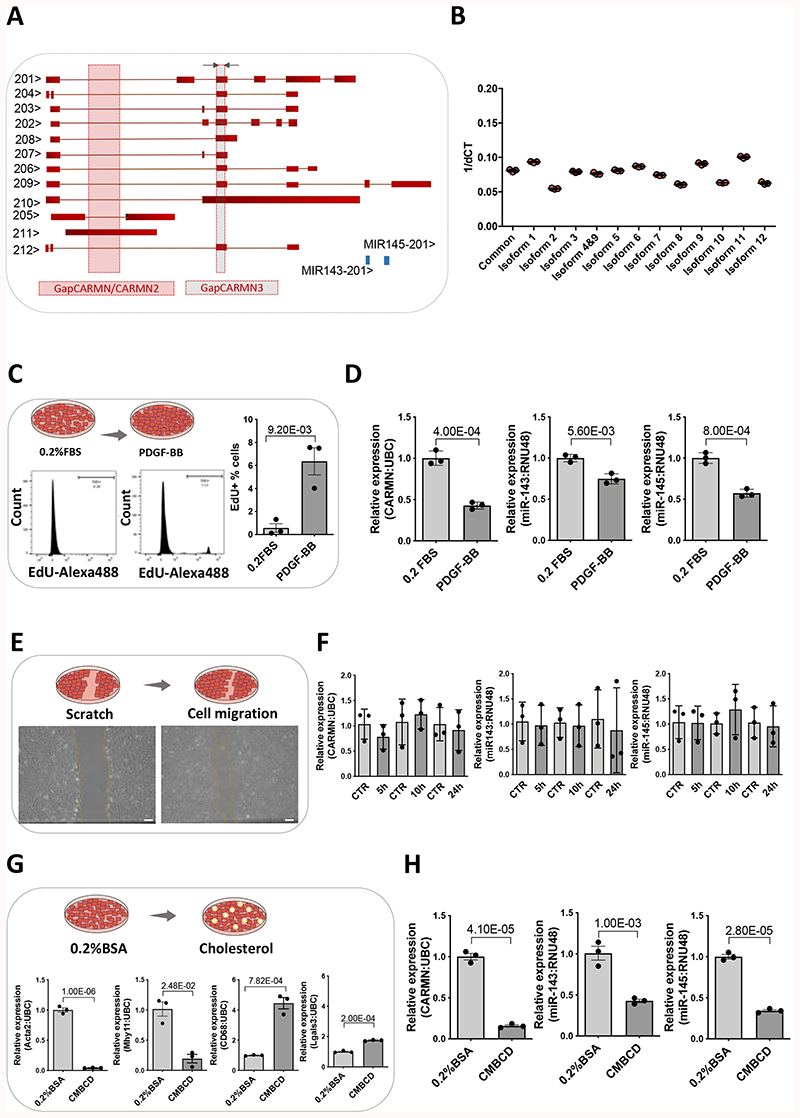
Figure 1. CARMN expression in basal and stimulated human primary CASMCs.
A) Schematic representation of human CARMN splice variants and pre-miRNAs miR-143 and miR-145 based on ENSEMBL version (GRCh38/hg38), including the position of the forward (Fw) and reverse (Rev) primers used to amplify all transcripts, grey arrows (except 4, 10 and 11) and the target region for the GapmeRs, GapCARMN, GapCARMN2 and GapCARMN3. The black arrows next to each transcript name, indicate the direction of transcription. B) Expression levels of CARMN transcripts in hCASMCs (n=3) via qRT-PCR using isoform-specific primers (Isoform 1,2,3,4+9,5,6,7,8,9,10,11, and 12) and common primers (amplifying all isoforms except 4, 10 and 11). C) Diagram of hCASMCs in basal condition (0.2% FBS) and treated for 48 hours with PDGF-BB. Histogram plots obtained by FACS showing the percentage of hCASMCs (n=3) up-tacking EdU following PDGF-BB treatment and in basal (0.2% FBS) conditions. Student’s t-test was used to assess statistical significance indicated with p values. D) QRT-PCR to show expression of CARMN, miR-143 and miR-145 in hCASMCs (n=3) treated with PDGF-BB. Student’s t-test was used to assess statistical significance indicated with p values. E) Diagram of the wound healing assay performed in hCASMCs (n=3), representative micrographs of hCASMCs scratch assay and migrating cells (Scale bar 100μm) and qRT-PCR showing the expression of CARMN and miR-143/145 G) Diagram of hCASMCs (n=3) in basal condition (0.2% BSA) and treated with cholesterol-methyl-b-cyclodextrin treated for 72 hours and expression levels of dedifferentiation markers (ACTA2, MYH11, CD68 and Lgals3), CARMN and miR-143/145 by RT-qPCR. Student’s t-test was used to assess statistical significance indicated with p values. H) qRT-PCR data showing the expression of CARMN and miR-143/145 in basal (0.2% FBS) and following CMBCD-loading treatment. Student’s t-test was used to assess statistical significance indicated with p values.