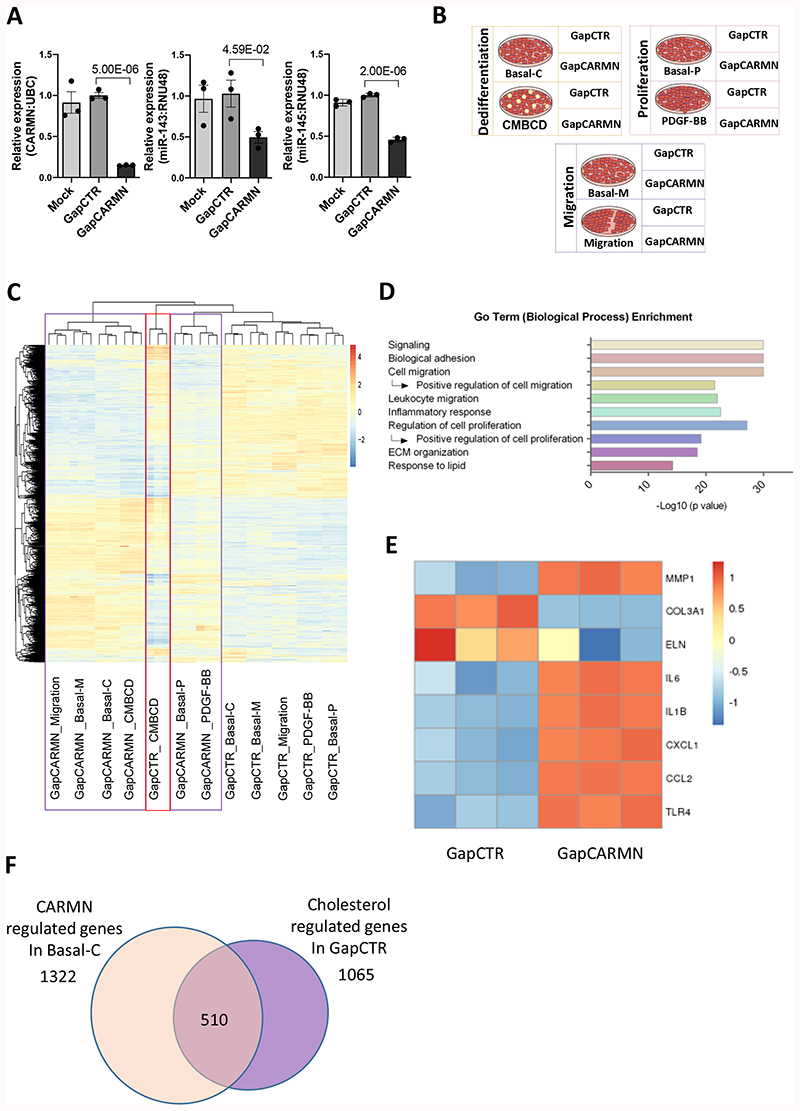
Figure 2. CARMN-regulated genes relevant to vSMC proliferation, migration and water-soluble cholesterol dependent dedifferentiation.
A) QRT-PCR results showing the expression of CARMN, miR-143 and miR145 hCASMCs (n=3) following transfection with GapmeR targeting CARMN (GapCARMN) and GapmeR control (GapCTR). Mock indicates un-transfected cells. One-way ANOVA with Bonferroni multiple comparison test was used to assess statistical significance indicated with p values. B) Schematic representation of the RNA-seq study design. C) Heatmap (as z-score of log2(FPKM+1)) of differentially regulated genes by CARMN depletion in all conditions. The purple box highlights the clustering of all CARMN-knock down samples while the red box shows the CMBCD treated replicates (Cont_KD). D) Graph of selected Go Term (Biological Process). The analysis was performed on the 2315 genes regulated by CARMN depletion in any conditions. E) Heatmap (as z-score of log2(FPKM+1)) of selected differentially regulated genes by GapCARMN and GapCTR in the basal condition with the shortest time of GapmeR transfection (migration). F) Venn diagram showing the overlap between genes regulated by CARMN depletion (in Basal-C condition) and genes regulated by CMBCD treatment (in GapCTR condition) in the same direction.