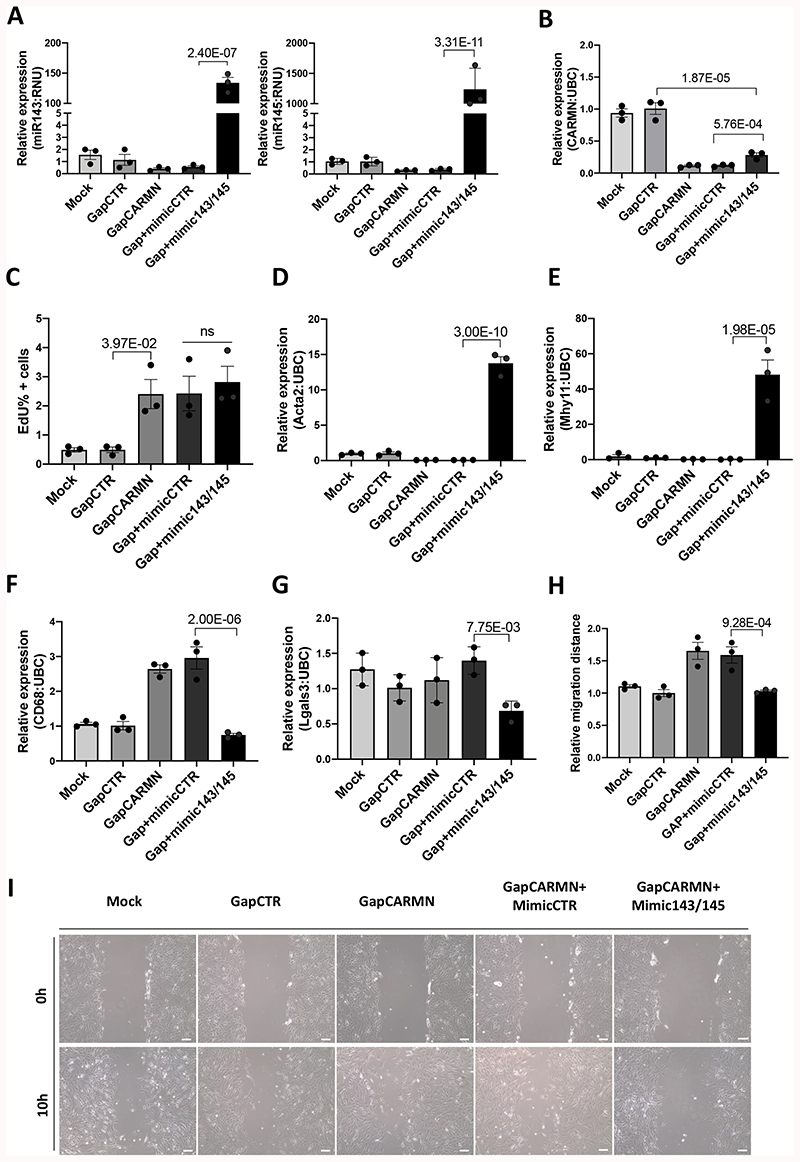
Figure 4. CARMN depletion regulated hCASMCs phenotypes in miRNAs-dependent and independent mode.
A), B) QRT-PCR expression data of miR-143, miR-145 and CARMN relative to housekeeper gene (RNU and UBC respectively) in GapCARMN, control (GapCTR), a combination of GapCARMN and mimic control (Gap+mimicCTR) and a combination of GapCARMN and mimic overexpressing miR143 and miR145 (GapCARMN+mimic143/145). Mock refers to un-transfected control cells. One-way ANOVA with Bonferroni multiple comparison test was used to assess statistical significance indicated with p values. C) Percentage of EdU positive hCASMCs (n=3) evaluated by FACS analysis and analyzed using FlowJo software. One-way ANOVA with Bonferroni multiple comparison test was used to assess statistical significance indicated with p values. D), E), F), G) qRT-PCR expression data relative to Acta2, Myh11, CD68 and Lgals3, respectively relative to housekeeper gene UBC in the above-mentioned transfection conditions. One-way ANOVA with Bonferroni multiple comparison test was used to assess statistical significance indicated with p values. H), I) Quantification of the relative migration distances of hCASMCs (n=3), obtained using ImageJ tool and representative images of hCASMCs at 0 and 10 hours from the induction of the scratch. Images acquired at 10X magnification, scale bar 100μm. One-way ANOVA with Bonferroni multiple comparison test was used to assess statistical significance indicated with p values. ns= not significant.