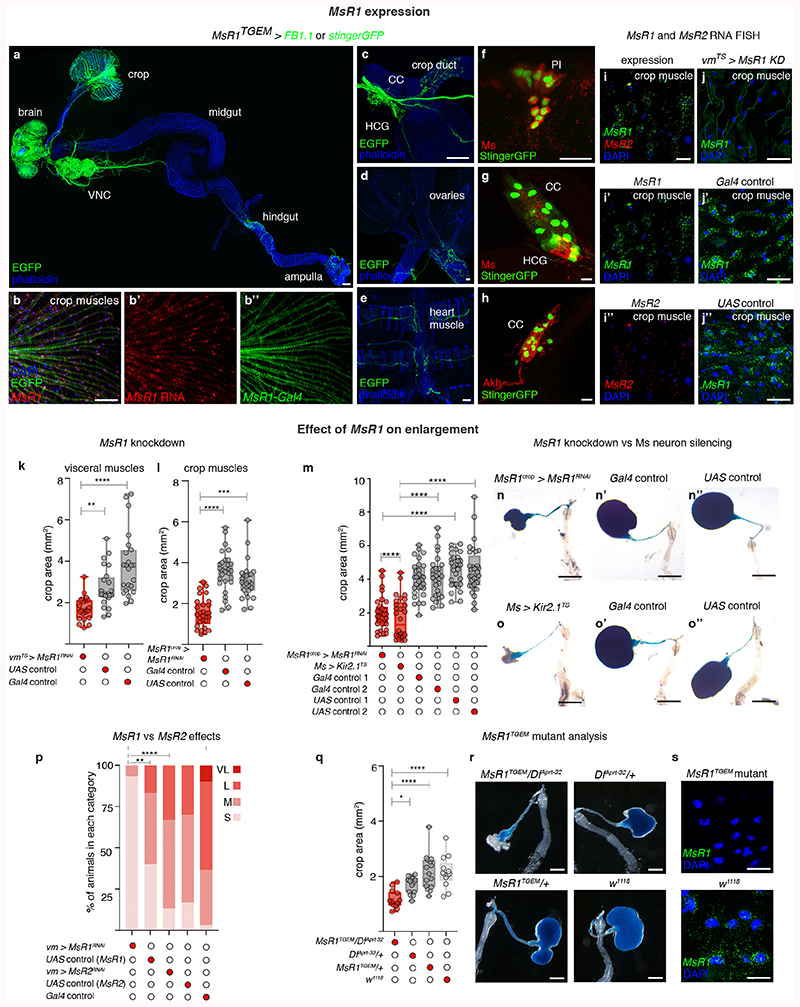
Extended Data Fig. 5. Expression of Ms receptors and their regulation of crop enlargement.
a, FB1.1-derived EGFP reveals MsR1 expression in the crop muscles and nervous system, including nerves innervating the crop, hindgut and rectal ampulla. In this and subsequent panels, muscles are labelled with phalloidin (in blue). b-b″, Co-expression between MsR1 mRNA stained with single-molecule RNA fluorescence in situ hybridisation (b,b’, in red) and FB1.1-derived EGFP driven by MsR1TGEM-Gal4 (b,b″, in green) is observed in crop muscles. Muscle nuclei are shown in blue with DAPI; single channels are shown for clarity. c, Detail of the HCG and corpora cardiaca (CC); the latter is extensively innervated by MsR1-expressing neurons. d, FB1.1-derived EGFP reveals MsR1 expression in neurons innervating the female reproductive system, but not in its muscles. e, FB1.1-derived EGFP reveals MsR1 expression in heart-innervating neurons, but not in heart muscles. f, Higher magnification image of the central brain; nuclear GFP reveals broad MsR1 expression in neurons including the PI Ms neurons shown with Ms staining (in red). g, A subset of 2-3 MsR1-positive neurons in the HCG co-express Ms. h, Nuclear GFP reveals MsR1 expression overlaps with Akh staining in CC cells (in red). i, Single-molecule fluorescence in situ hybridisation of MsR1 and MsR2 mRNAs in crop muscles; MsR1 (in green) is more readily detected than MsR2 (in red). Muscle cell nuclei are shown in blue by DAPI staining. The MsR1 expression described in a-h is consistent with transcriptomics data88,89. i’ and i″ show single MsR1 or MsR2 channels for clarity. j-j’, Validation of adult-specific MsR1 knockdown in visceral muscles (vmTS > MsR1-RNAi). Panels show high magnification images of crop muscles. MsR1 mRNA expression is visualised by single-molecule RNA fluorescence in situ hybridisation (in green) in vm-Gal4TS (j), but it is reduced/absent from MsR1 knockdown crops (j’). k, Quantifications of crop area in starved-refed flies upon downregulation of MsR1 in visceral muscles, showing that crop size is visibly reduced upon MsR1 downregulation compared to UAS and Gal4 controls. l, A similar reduction in crop area is also quantified upon MsR1 downregulation specifically in crop muscles using a different driver line (MsR1crop > MsR1RNAi). MsR1crop-Gal4 is MsR1-Gal4, nsyb-Gal80, in which MsR1-Gal4 neuronal expression is prevented using the pan-neuronal nsyb-Gal80 driver, rendering it a crop muscle-specific driver. m-o″, Effect of crop muscle-specific downregulation of MsR1 on crop size. m, Quantifications of crop area in starved-refed mated females shows that crop-specific downregulation of MsR1 (MsR1crop > MsR1RNAi) resulted in reduced crop areas, similar to Ms neuron inactivation (Ms > Kir2.1) and significantly reduced as compared to Gal4 and UAS controls. n-o″, Representative crop phenotypes of the genotypes quantified in m. p, Quantification of crop area upon visceral muscle-specific MsR1 and MsR2 downregulation, showing that MsR1 knockdown, but not MsR2 knockdown, resulted in reduced crop sizes, as compared to UAS and Gal4 respective controls. q, Quantifications of crop area in starved-refed mated females shows that heteroallelic MsR1TGEM/DfAprt-32 mutants have reduced crop areas relative to w1118 or heterozygous controls. r, Representative crop images from genotypes quantified in q. s, Validation of MsR1 mutation and MsR1 fluorescence in situ hybridisation signal specificity. MsR1 mRNA (green) is absent from the crop muscle cells of MsR1TGEM mutants, and apparent in w1118 control flies. Scale bars: b-b″, f-j″ and s = 10μm, a, c, d, e = 50μm, r = 500μm and n-o″ = 1mm. See Supplementary Information for a list of full genotypes, sample sizes and conditions. In all boxplots, line: median; box: 75th-25th percentiles; whiskers: minimum and maximum. All data points are shown. *: 0.05>p>0.01; **: 0.01>p>0.001; ***: p<0.001.