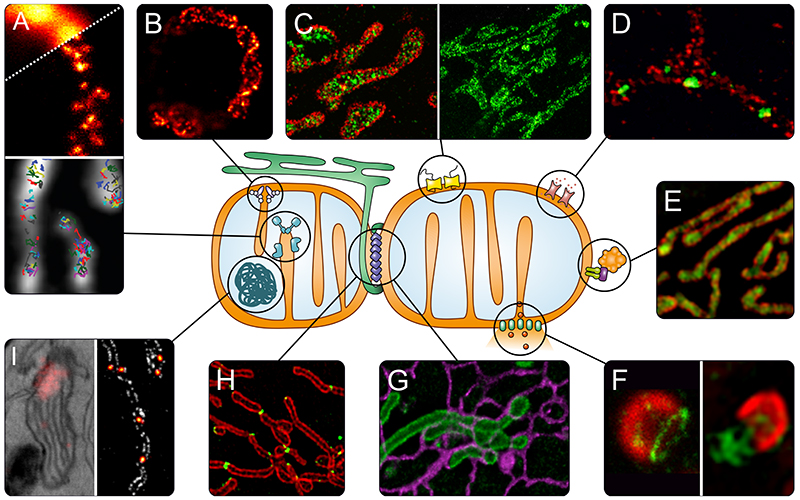
Fig. 2. Super-resolution microscopy to address questions of mitochondrial biology.
A) OXPHOS. Upper panel: Distribution of assembly factors recorded with STED nanoscopy (80). Lower panel: Tracking of single OXPHOS subunits using SMLM (77). B) Localization of the MICOS subunit Mic60 in a yeast cell, revealed by STED nanoscopy (89). C) TOM complex in the outer membrane recorded with DNA-PAINT nanoscopy (left, TOM complex in red) (63) and STED nanoscopy (right) (67). D) Differential distributions of the three human hVDAC (mitochondrial porin) isoforms in the outer membrane of human mitochondria (STED nanoscopy, hVDAC in green) (66). E) Sub-mitochondrial localization of PINK1 (green) shown by 3D-SIM (76). F) Apoptosis. Left: Several pro-apoptotic BAX proteins (green) form ring like structures in the outer membrane that may act as pores (STED nanoscopy) (100). Right: During later steps of apoptosis, these large BAX assemblies facilitate the herniation of the inner membrane and release of mtDNA (green) as shown by SIM (102). G) Interactions between mitochondria (green) and the ER (purple) in living cells recorded by STED nanoscopy (70). H) Spatial dynamics of the dynamin-like GTPase DRP1 (green), which is essential for mitochondrial fission, visualized by SIM (109). I) Nucleoids, analyzed with correlative PALM and EM (left panel) (131), and with STED nanoscopy (right panel, nucleoids in fire) (unpublished).