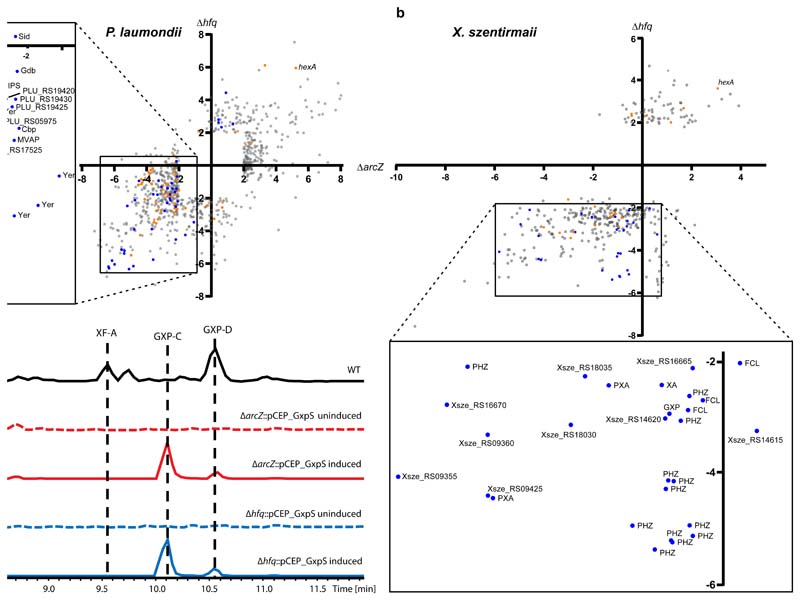
Figure 4.
Comparison of ArcZ and Hfq regulon a. P. laumondii and b. X. szentirmaii. Scatterplots show individual coding sequences and their corresponding regulatory changes compared to wild type in either the ΔarcZ (x-axis) or Δhfq (y-axis) mutants, with SMs (blue dots) and regulators (orange dots) highlighted. The inset shows only SM-related coding sequences, including those associated with anthraquinone (AQ), mevalagmapeptide (MVAP), carbapenem (Cbp), yersiniabactin (YER), GameXPeptide (GXP), siderophore (SID), isopropylstilbene (IPS) and glidobactin (Gdb), phenazine (PHZ), fabclavine (FCL), xenoamicin (XA) and pyrrolizixenamide (PXA). C Base peak chromatograms (BPCs) of X. szentirmaii WT (black), ΔarcZ::pCEP_GxpS uninduced (red dotted line), ΔarcZ::pCEP_GxpS induced (red solid line), Δhfq::pCEP_GxpS uninduced (blue dotted line) and Δhfq::pCEP_GxpS induced (blue solid line). Peaks corresponding to (cyclo)tetrahydroxybutyrate (THB56), Linear GameXPeptide C (GXP-C), rhabdopeptide 772 (RXP), xenofuranone A (XF-A), as well as cyclic GXP-C and GXP-D. Five times zoom was applied to base peak chromatograms in the uninduced and wild type samples.