Figure 4. The structure of the in situ hook cap complex (FlgD).
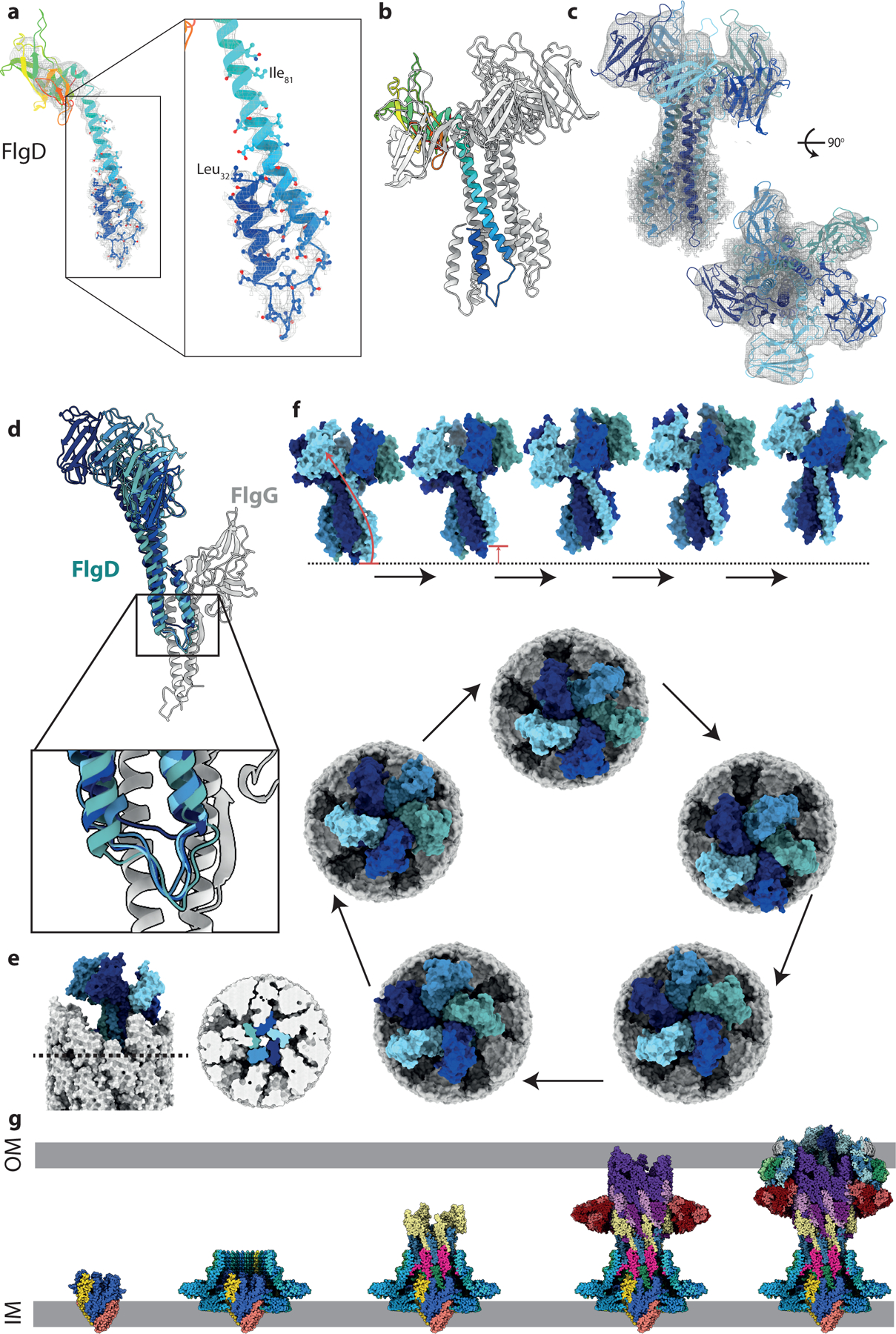
a, The N-terminus of FlgD, the hook cap protein forms an unexpected bent-back helical pair. b, Pentamer of FlgD, with one copy represented as a rainbow. c, FlgD pentamer, colored by chain, shown in the experimental local resolution volume (grey mesh). (d) Overlay of five FlgD (blue) – FlgG (grey) pairs showing the main interface. One copy of FlgD (navy blue) sits particularly high against its cognate FlgG, showing the FlgD helices can slip vertically with respect to FlgG. e, Surface representation of the tip of the axial components with the cap in situ shows that the top sits at an angle to the rod axis. Right hand panel shows a cut-through at the level indicated in the left-hand panel allowing visualisation of the tight packing of the FlgD blades created by the paired helices at the N-terminus. (f) Proposed motions involved in FlgD cap catalysed assembly mechanism. Top panel (Supplementary Movie 1) show a side view of four sequential cap movements orientated as in part (e), with the rod removed for clarity. In the first transition, the movement of the lowest copy (light blue) to become the highest copy is illustrated with an arrow. The bottom panel shows a top down view of the cap on the rod, showing a full revolution of the cap around the rod, sequentially opening up rod/hook subunit binding sites, without any rotation of the cap structure on its axis. (g) Assembly steps of the flagellar basal body shown using the sub-structure solved in this study. Export gate nucleates correct assembly of the MS-ring and seeds the protofilaments of the rod. Rod growth to the point of FlgG allows assembly of the P-ring around the IM-proximal end of the distal rod, which can then act as an assembly point for the L-ring. OM-tethering of FlgH places the LP-ring at an appropriate height on the distal rod. Finally, FlgD assembly on the tip of the distal rod permits correct insertion of hook subunits via stepped revolution.
