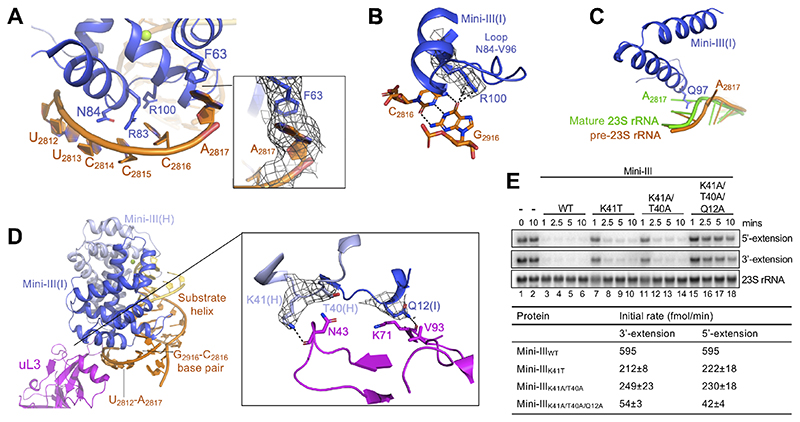
Figure 4. Interactions between Mini-III and 50S(pre-23S).
(A) Mini-III interactions outside of the substrate helix. Conserved residues are shown in stick representation. Insert shows the cryoEM density for the flipped-out A2817 that forms a π-π stacking interaction with F63(I).
(B) CryoEM density for Mini-III residue R100(I) bound to the C2816-G2916 base-pair from pre-23S rRNA, coloured as in Figure 1A.
(C) Superimposition of mature 23S rRNA (green, PDB 5NJT) with Mini-III-bound pre-23S rRNA (orange). Mini-III residue Q97(I) (dark blue, stick representation) sterically clashes with A2817 in mature 23S rRNA, but not in pre-23S rRNA.
(D) Mini-III (dark/light blue), uL3 (pink), and pre-23S rRNA (orange/yellow) interactions. RNA elements shown in (A), (B), and (C) are highlighted. The insert shows cryoEM density for residues from Mini-III (dark/light blue) able to bind to uL3 (pink).
(E) Top: Northern blot showing removal of 5’-and 3’-extensions from pre-23S rRNA by Mini-IIIWT, Mini-III-K41T, Mini-III-K41A/T40A, and Mini-III-K41A/T40A/Q12A. The first two lanes show pre-23S rRNA at 0 and 10 minutes in the absence of Mini-III protein. The blot is representative of at least 3 independent experiments. Bottom: Initial reaction rates for each mutant. Initial slopes (%/min) were calculated for each curve by regression of the linear portion of the curves shown in Figure S2D, with 100% corresponding to 700 fmol pre-23S rRNA.