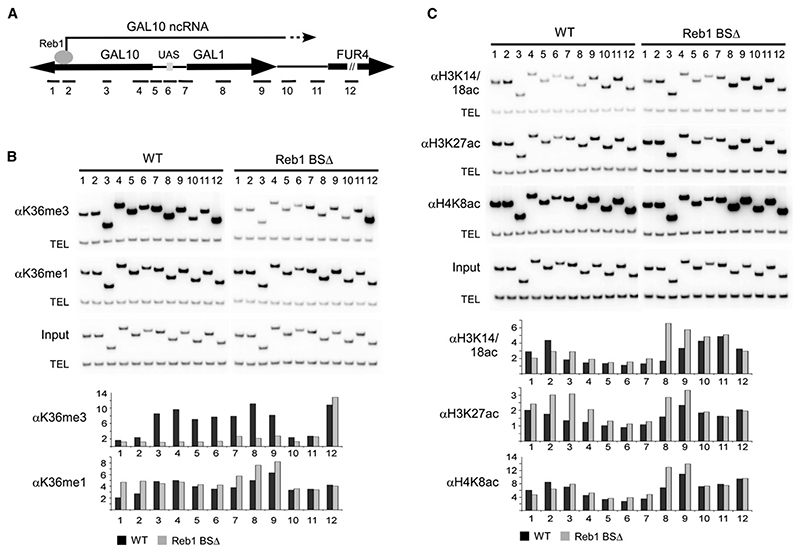
Figure 4. Histone Modifications Induced by the GAL10 ncRNA.
(A) Schematic of the GAL locus showing the location of primer sets used for ChIP analysis.
(B) ChIP analysis of H3 K36 methylation in the presence and absence of the GAL10 ncRNA (Reb1 BS+/−). Cells were grown to log phase in rich media containing 2% raffinose, and chromatin was precipitated with antibodies against H3 K36me3 and H3 K36me1. PCR was performed with primer sets shown on the schematic diagram for the GAL locus and a telomeric primer set (TEL) as a loading control.
(C) ChIP analysis of histone acetylation in the presence and absence of the GAL10 ncRNA (Reb1 BS+/−). Cells were grown to log phase in rich media containing 2% raffinose, and chromatin was precipitated with antibodies against acetylated H3 K14/18, H3 K27, and H4 K8. PCR was performed with primer sets shown on schematic diagram for the GAL locus and a telomeric primer set (TEL) as a loading control.
Graphs in (B) and (C) show quantification of ChIP data as fold enrichment of signal at the GAL locus over telomeric signal normalized to the input. Data shown is the average of two independent experiments.