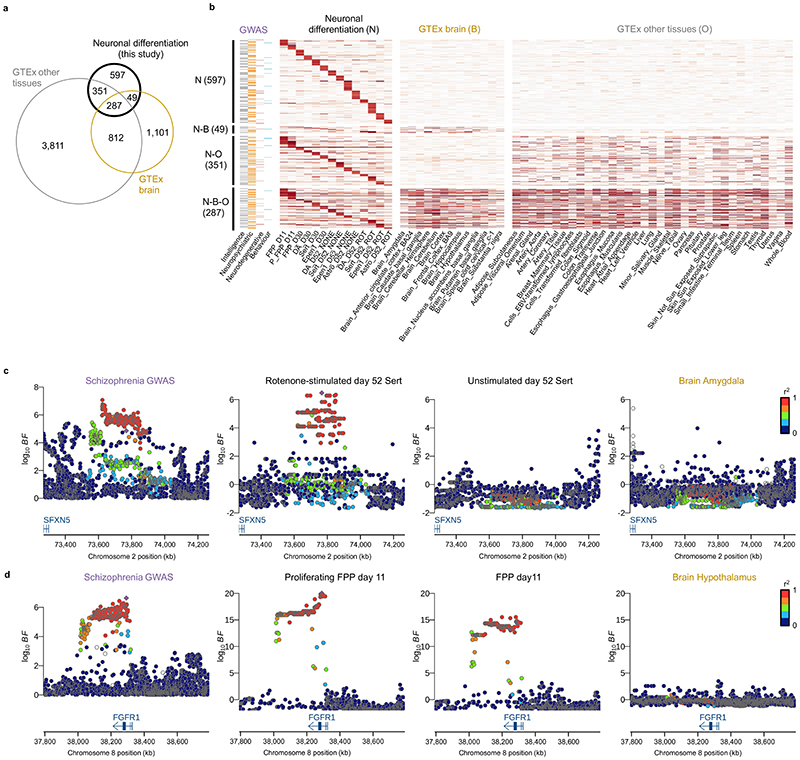
Fig. 5. Colocalization analysis of eQTL with 25 neuro-related GWAS traits.
(a) Venn diagram showing the number and the overlap of colocalization events discovered using eQTL maps from this study, GTEx brain and other GTEx tissues. (b) Heatmap showing the posterior probability of colocalization (PP4 from COLOC; Methods) for eQTL that colocalized with one or more GWAS traits. N: Neuronal differentiation (this study), B: GTEx Brain, O: Other GTEx tissues. (c) Locus zoom plots around the SFXN5 gene. The schizophrenia GWAS association (left) is colocalized with the eQTL in rotenone-stimulated serotonergic-like neurons at D52 (second panel from the left). No colocalization signal was detected in unstimulated serotonergic neurons at D52 (third panel from the left) or any other brain GTEx tissues as illustrated here with GTEx brain amygdala (rightmost panel). The lead variant is indicated with a purple diamond and other points were colored according to the LD index (r2 value) with the lead variant. (d) A midbrain progenitor-specific eQTL for FGR1 associated with schizophrenia. We identified a colocalization event with this eQTL in both proliferating (second panel from the left) and non-proliferating floor plate progenitors (third panel from the left) at day 11. No colocalization was found in any other cell type profiled in this study (not shown) nor in any brain GTEx tissues (shown with GTEx brain hypothalamus, rightmost panel). Astro: Astrocyte-like; DA: Dopaminergic neurons, Epen1: Ependymal-like1, FPP: Floor Plate Progenitors, P_FPP: Proliferating FPP, Sert: Serotonergic-like neurons, U_Neur1,2,3: Unknown neurons 1,2,3.