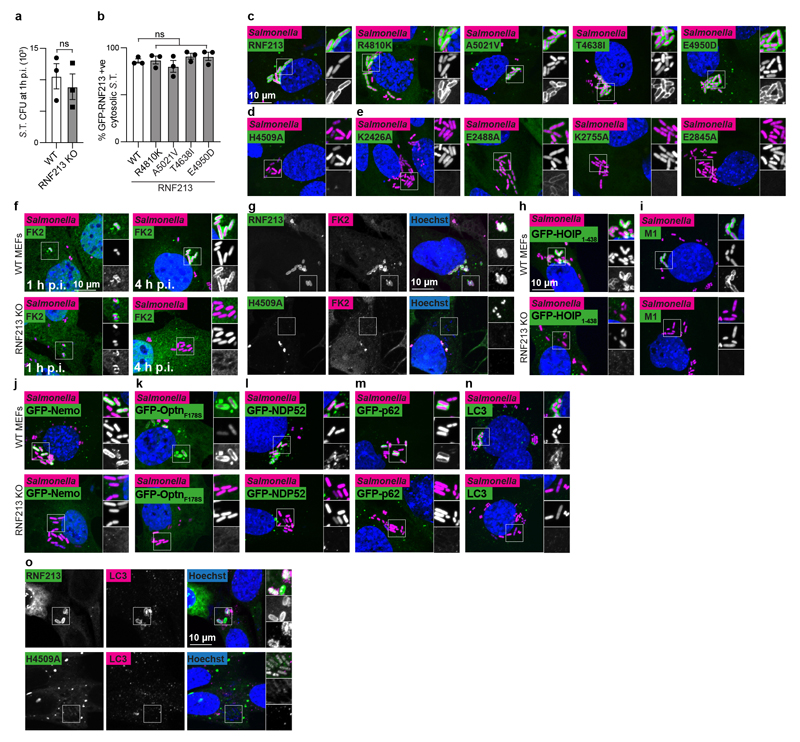
Extended Data Figure 4. RNF213 provides cell-autonomous immunity.
a, S. Typhimurium CFU 1 h p.i. extracted from WT and RNF213KO MEFs. Bacteria were counted by serial dilution of cell lysate on LB agar plates. Data are expressed as the mean ±s.e.m. of three experiments. b, Percentage of cytosolic S. Typhimurium positive for FLAG-GFP-RNF213 at 3 h p.i. in RNF213KO MEFs stably expressing the indicated GFP- RNF213 alleles. c-o, representative confocal micrographs for Fig.4d–o. c-e, WT or RNF213KO MEFs stably expressing the indicated GFP-RNF213 alleles infected with mCherry-expressing S. Typhimurium, fixed 3 h p.i. f-o, WT and RNF213KO MEFs (f,g,n), RNF213KO MEFs stably expressing GFP-RNF213 H4509A (g,o) or WT and RNF213KO MEFs stably expressing GFP- HOIP1-438 WT or T360A (h), GFP-Nemo (j), GFP-OptineurinF178S (k), GFP-NDP52 (l), GFP- p62 (m), infected with mCherry-expressing (c-f, h-n) or BFP-expressing (g,o) S. Typhimurium, fixed 3 h p.i. and stained for FK2 (ubiquitin) (f,g), M1-linked linear ubiquitin chains (i) or LC3 (n,o). g,o, For better visibility of bacteria, Hoechst/BFP channel has been depicted in white in the zoomed sections. c-n, Scale bar, 10 pm. Statistical significance was assessed by twotailed unpaired Student’s t-test (a) or one-way anova (b). ns, not significant. Data are expressed as the mean ± s.e.m. of 3 independent experiments (a,b, source data Extended Data Fig.4) and micrographs are representative of 3 biological repeats (c-o)