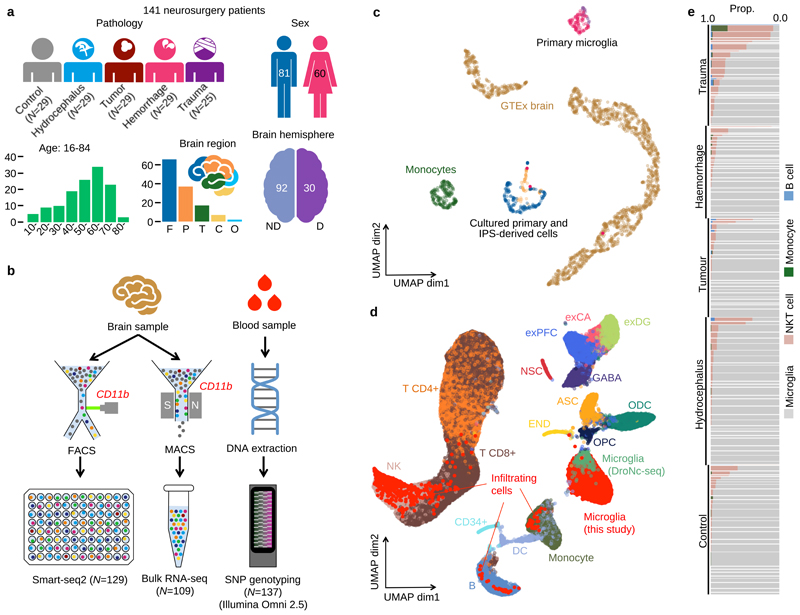
Figure 1. Study design and overview of the data.
a. Metadata from 141 neurosurgery patients enrolled in this study. Brain region annotation: Cerebellum (C); Frontal (F); Occipital (O); Parietal (P); Temporal (T); non-dominant (ND); dominant (D). b. Experimental design using Smart-seq2 and bulk RNA-seq with SNP genotyping. c. UMAP of bulk RNA-seq from myeloid cells and brain tissue. The “Primary microglia” cluster contains samples collected in this study (pink dots) and previous studies (purple dots) (information on the source of previous study data can be found in Supplementary Table 7). “Cultured primary and IPS-derived cells”, includes IPS-derived macrophages and microglia (blue dots), cultured primary microglia and monocyte derived macrophages (orange dots). “Monocytes” (green dots) denotes primary monocytes obtained from the BLUEPRINT project, and “GTEx brain” denotes all brain tissues from GTEx v7. The left cluster of GTEx brain corresponds to cerebellum or cerebellar hemisphere samples and the right cluster contains samples from all other brain regions. d. UMAP of single-cell RNA-seq data combined with 68K PBMC scRNA-seq18 and whole brain DroNc-seq19. Bright red dots represent cells collected in this study. Cell type annotations were obtained from: glutamatergic neurons from the PFC (exPFC); pyramidal neurons from the hip CA region (exCA); GABAergic interneurons (GABA); granule neurons from the hip dentate gyrus region (exDG); astrocytes (ASC); oligodendrocytes (ODC); oligodendrocyte precursor cells (OPC); neuronal stem cells (NSC); endothelial cells (END); dendritic cell (DC); B cell (B); hematopoietic progenitor cell (CD34+); NK T cell (NK). e. Proportions of non microglia for each patient in our data. Each horizontal bar corresponds to one patient. The thickness of each bar is proportional to the number of cells observed for the patient. Patients are stratified by pathology.