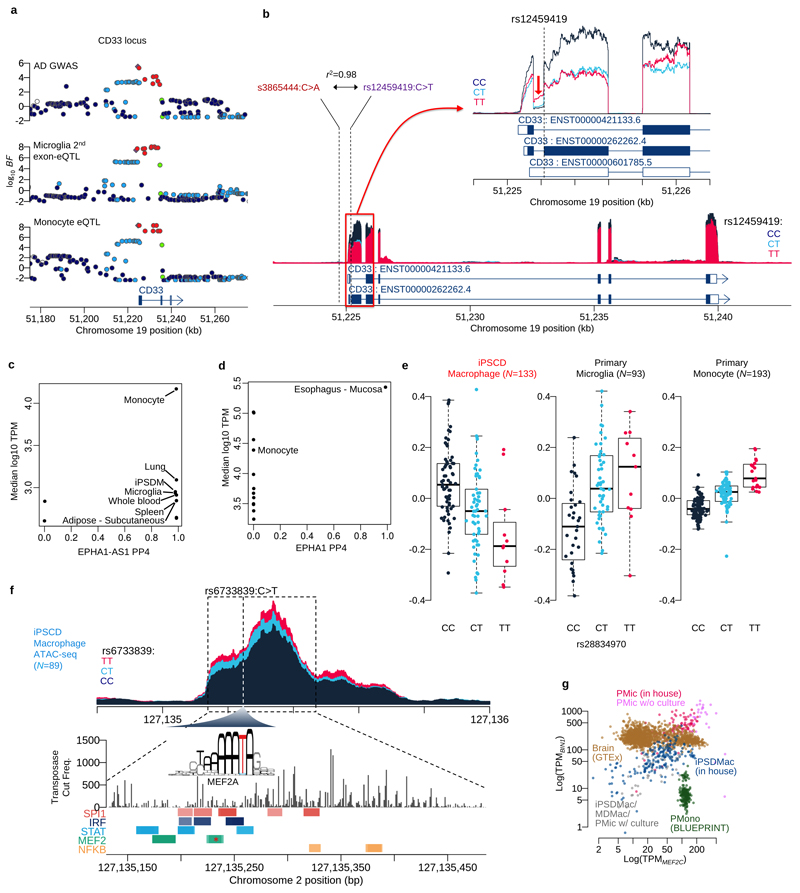
Extended Data Fig. 5. Finemapping of microglia eQTLs.
a. Regional association plots at the CD33 locus. b. Coverage plot shows the normalised expression level around the CD33 gene stratified by genotype at the putative splice variant (rs12459419C>T). The zoom-in panel shows a coverage plot of expression level around the second exon (ENST0000262262.4). The coverage shows the first intron expression is negatively correlated with the second exon expression, suggesting the expression of non-coding isoform (ENST00000601785.5) is increased by the alternative allele (T) of the splicing QTL. c. Colocalisation between an association with risk for Alzheimer’s disease on chromosome 2 and an eQTL for the noncoding RNA gene EPHA1-AS1 in microglia, GTEx tissues and myeloid cell types. The x-axis shows the posterior probability of colocalisation (PP4) and y-axis shows the average expression level (log10 TPM) for each tissue or cell type. d. Colocalisation between AD risk and expression of the protein-coding EPHA1 gene. The x-axis shows the posterior probability of colocalisation (PP4) and y-axis shows the average expression level (log10 TPM) for each tissue or cell type. e. Boxplots show the relationship between expression at the PTK2B gene and genotype at the lead eQTL variant (rs28834970C>T) three myeloid cell types. The y-axis shows normalised expression levels (log TPM value). Each dot on the box shows the expression level of a single sample. f. Coverage plot shows chromatin accessibility in iPS cell derived macrophages stratified by three genotype groups of the lead AD GWAS/BIN1 eQTL variant g. Scatter plot of MEF2C (x-axis) and BIN1 (y-axis) expression in GTEx brain tissues and myeloid cell type.