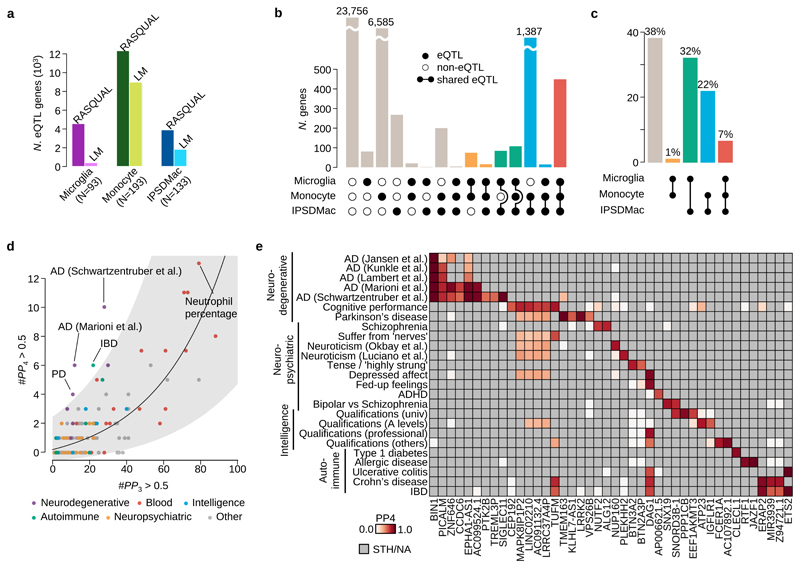
Figure 4. Mapping and colocalisation of microglia eQTLs with various GWAS traits.
a. The numbers of eQTL genes discovered by two different methods, RASQUAL (left bar) or simple linear regression (right bar, LM) in three myeloid cell types at FDR 5% (see Online Methods). b. The number of shared eQTLs across three myeloid cell types obtained by the three-way Bayesian hierarchical model (Online Methods). The combination of genes that are eQTLs (closed dots) or non-eQTLs (open dots) across three different myeloid cell types are shown below each bar. A line connecting two dots indicates a shared eQTL between different cell types. c. Empirical prior probability of eQTL sharing among three different myeloid cell types obtained by the three-way Bayesian hierarchical model (Online Methods). The Y-axis shows the proportion of genes genome-wide and the dots connected by segment illustrate the shared genetic association. d. Colocalisation of microglia eQTLs with 146 GWAS traits. The x-axis shows the number of genes where PP3, the posterior probability of the microglia eQTL and GWAS association being driven by two independent causal variants, was greater than 0.5. The y-axis is the number of colocalised genes where the posterior probability of a single shared causal variant between a microglia eQTL and a GWAS locus (PP4) was greater than 0.5. We subdivided and colored GWAS traits as follows: purple: neurodegenerative diseases; red: blood cell trait; blue: traits related with intelligence; green: autoimmune diseases; yellow: neuropsychiatric diseases; gray: others. The line shows a log-normal linear regression fit with gray shaded area indicating the 95% prediction interval of the fit. e. Heatmap of PP4 for neuro-degenerative/psychiatric diseases, intelligence related traits and autoimmune diseases showing all genes and GWAS trait with a combined PP4 greater than 0.5. Gray cells indicate that the gene-trait combination was not tested because the GWAS locus was not significant (lead SNP P>10-6), or there were no GWAS summary statistics available for secondary hits (PTK2B and TREML3P).