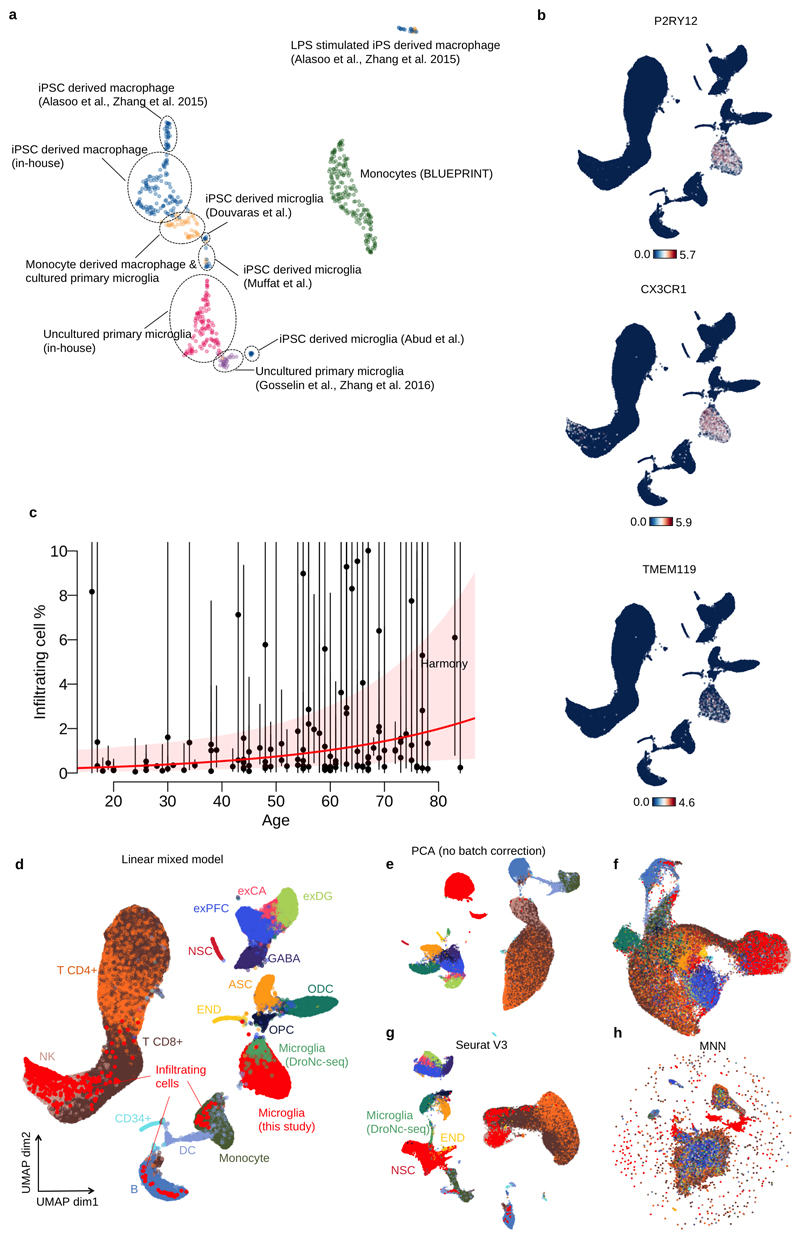
Extended Data Fig. 1. Overview of bulk and single cell RNA-seq data.
UMAP of bulk RNA-seq for myeloid cells. The “Primary microglia” cluster contains samples collected in this study (pink dots) and previous studies (purple dots) (information on the source of previous study data can be found in Supplementary Table 7). “Cultured primary and IPS-derived cells”, includes IPS-derived macrophages and microglia (blue dots), cultured primary microglia and monocyte derived macrophages (orange dots). “Monocytes” (green dots) denotes primary monocytes obtained from the BLUEPRINT project. b. Feature plots of three microglia marker genes (P2RY12, CX3CR1 and TMEM119) using the same UMAP coordinates as Figure. 1d. c. Age versus percentage of infiltrating cells. Red line shows the logistic regression line, the red transparent band shows the 95% confidence interval estimated using a generalised linear mixed model for the binary outcome (Materials and Methods). d. UMAP plot identical to Figure 1d. e. UMAP plot from the first 12 principal components computed from the same input data for the linear mixed model without any batch correction. f. UMAP of the same 12 PCs where the batch effect was corrected by using Harmony45. g. UMAP of batch corrected data using the canonical correlation analysis method implemented in Seurat V3 46 with a default setting. We computed the 12 PCs from the integrated data for UMAP plot. h. UMAP of batch corrected data using MNN correct47. Note that points were coloured according to the cell types (same as Figure 1d): glutamatergic neurons from the PFC (exPFC); pyramidal neurons from the hip CA region (exCA); GABAergic interneurons (GABA); granule neurons from the hip dentate gyrus region (exDG); astrocytes (ASC); oligodendrocytes (ODC); oligodendrocyte precursor cells (OPC); neuronal stem cells (NSC); endothelial cells (END); dendritic cell (DC); B cell (B); hematopoietic progenitor cell (CD34+); NK T cell (NK).