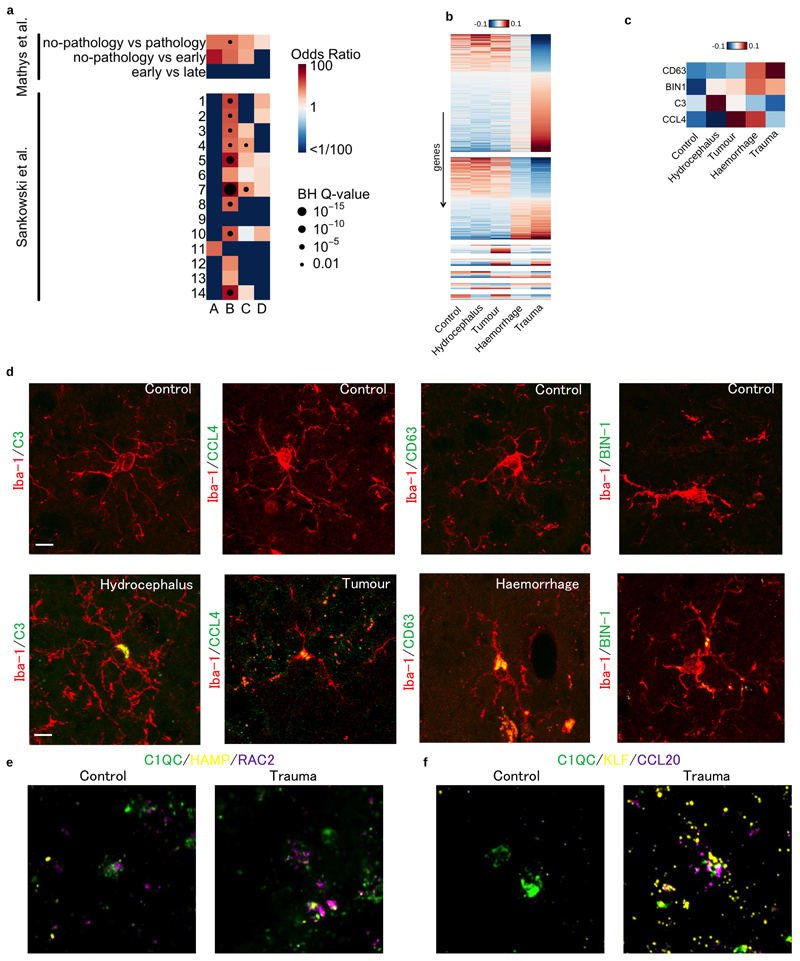
Extended Data Fig. 2. Microglia marker gene comparisons and validations.
a. Marker gene enrichment analysis with Alzheimer’s disease associated microglia20 and glioma associated microglia21. There are three different comparisons for Alzheimer’s disease associated microglia and 14 different populations for glioma associated microglia. Heatmap shows odds ratios and Benjamini-Hochberg (BH) Q-values of the Fisher exact tests between our marker genes and differentially expressed genes in other studies. b. Differentially expressed genes between microglia from different patient pathologies using single cell RNA-seq data. Heatmap shows averaged, normalised expression level (defined as the posterior mean of pathology random effect term, see Materials and Methods) of differentially expressed genes at local true sign rate (ltsr) greater than 0.9 ((Urbut et al. 2019); see Materials and Methods for details). Heatmap is divided into groups based on all possible pairwise groupings of the four cell populations, ordered by most transcriptionally distinct, such that the most different grouping, trauma versus all non-trauma, appears at the top. c. Differential expression of candidate marker genes for immunohistochemistry in fresh frozen patient tissue samples. d. Immunohistochemistry panel of each pathology to validate expression of a differentially expressed gene at the protein level; hydrocephalus (C3), tumour (CCL4), haemorrhage (CD63) and trauma (BIN-1) compared to control. Iba-1 (red) and protein of interest (green). e. RNAScope image of differentially expressed gene panel for cluster C; HAMP (yellow) and RAC2 (purple) with C1QC (green) used to identify microglia. f. RNAScope image of differentially expressed gene panel for cluster D; KLF (yellow) and CCL20 (purple). Scale bar 10μM.