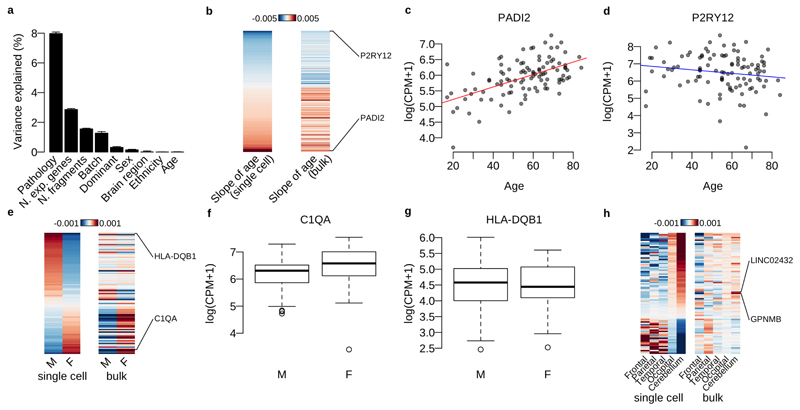
Extended Data Fig. 3. Differential expression analysis with bulk RNA-seq data.
a. Variance components analysis of log CPM values for the bulk RNA-seq data (N=102) with biological and technical factors using the linear mixed model (Online methods). b. Heatmap shows the effect size of age for each gene (each row) estimated by the linear mixed model (Online methods). The genes with LTSR>0.9 in single-cell data are shown. c. PADI2 normalised expression in bulk RNA-seq data against patients’ age. d. P2RY12 expression in bulk RNA-seq data against patients’ age. e. Heatmap shows the average expression of males and females for each gene (each row) estimated by the linear mixed model (Online methods). The genes with LTSR>0.9 in single-cell data are shown. f. C1QA normalised expression in bulk RNA-seq data for males (M) and females (F). g. HLA-DQB1 normalised expression in bulk RNA-seq data for males (M) and females (F). h. Heatmap shows the average expression for 5 different brain regions estimated by the linear mixed model (Online methods). The genes differentially expressed between a combination of Occipital and Cerebellum and the 3 other regions (LTSR>0.9) in single-cell data are shown.