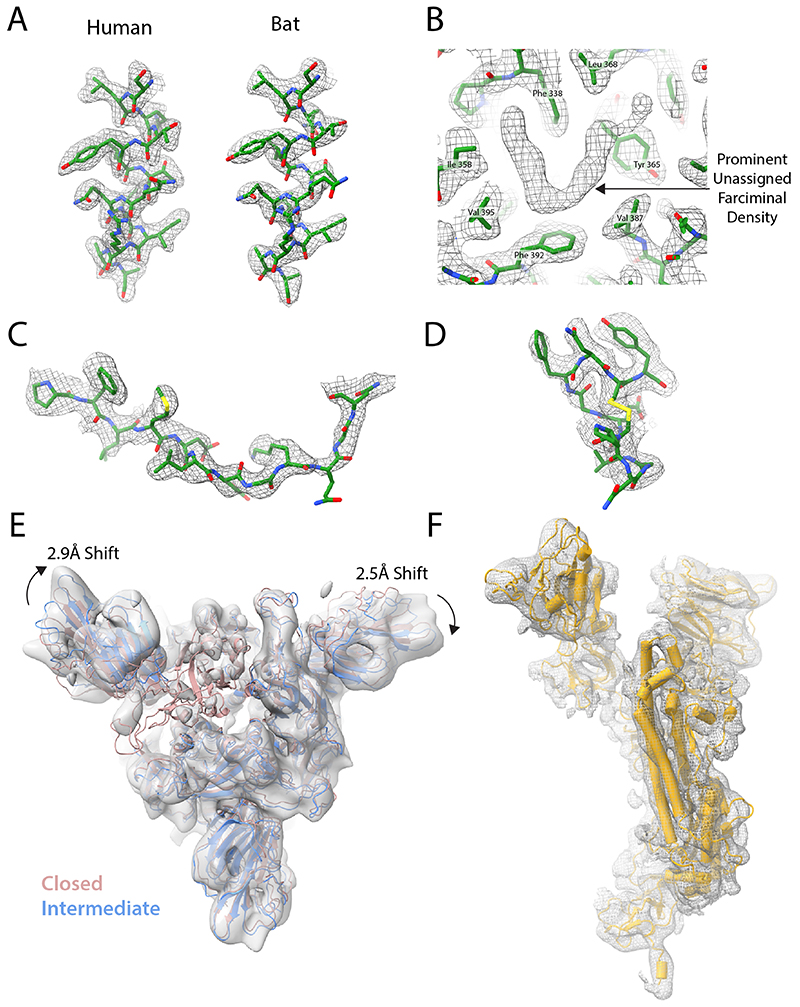
Extended Data Fig. 2. Density features of spike protein cryoEM density.
EM density shown as grey mesh, with built models shown in green. (A) Density for residues 1003-1016 of the uncleaved closed human and bat structures. (B) Unassigned farciminal density seen in maps of the closed conformation of human spike protein. (C+D) Typical density for examples of previously unbuilt external loops in uncleaved closed human structures: residues 174-185 of the NTD (C) and residues 479-489 of the RBD (D). (E) EM density of Intermediate conformation of furin-cleaved spike. The fitted model (blue) is compared to that of the closed conformation (pink). Shifts in the NTDs are indicated with arrows, with values calculated from translations of their centres of mass. (F) Example EM density of a monomer of the Intermediate conformation map with the built structure shown in yellow.