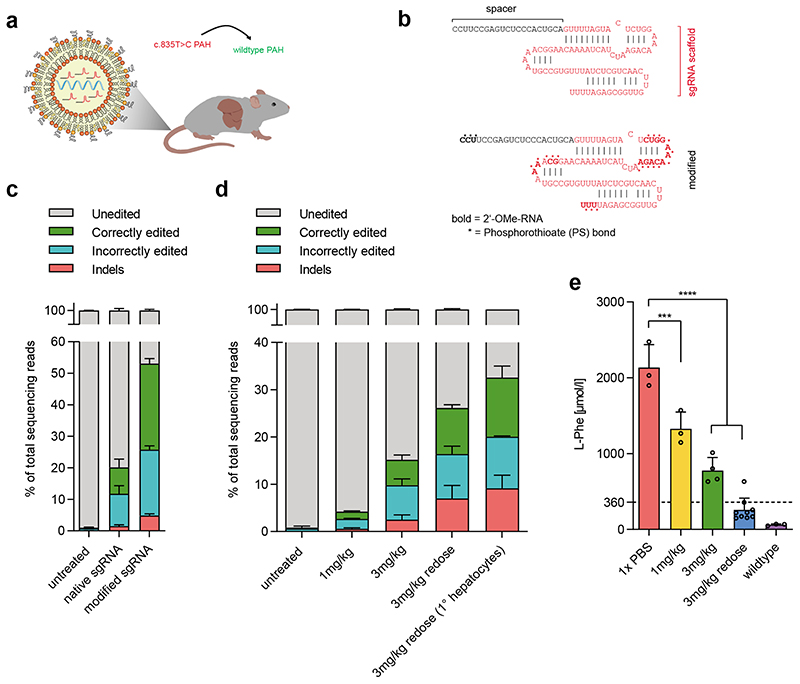
Figure 3. Correction of the disease locus of the Pahenu2 mouse model via LNP-mediated delivery of base editing components.
a) Concept of in-vivo correction of the mutated Pahenu2 locus by systemic administration of LNP-encapsulated SaKKH-CBE3 mRNA and sgRNA. b) Chemical modifications introduced to the Sa Cas9 sgRNA targeting the c.835T>C mutation. c) Correctly edited reads support restoration of the correct PAH amino acid sequence by C-to-T conversion of the target C13, including any synonymous mutations. Incorrectly edited reads summarize mutations that are not synonymous to the wildtype PAH amino acid sequence, including C-to-T conversions at positions additional to or other than C13, and conversions that are not C-to-T. Indels are shown separately. Values represent mean of three independent biological replicates performed on separate days ± s.d. d) Editing efficiencies after systemic administration of 1 mg/kg, 3 mg/kg or two doses of 3 mg/kg in vivo in whole liver lysates and in isolated primary hepatocytes. Correctly edited reads restore the wildtype PAH amino acid sequence, incorrectly edited reads mean mutations that lead to amino acid sequences nonsynonymous to the wildtype PAH amino acid sequence. Values represent mean of three independent biological replicates performed on separate days ± s.d. e) Blood L-Phe levels from homozygous Pahenu2 and wildtype C57BL6 mice were determined one week after the latest injection. Values (n=3 per treatment group) represent mean ± s.d. A two-way ANOVA with Dunnett’s multiple comparisons test was performed to account for multiple comparisons to a single control (untargeted): ***P < 0.0002, ****P < 0.0001.