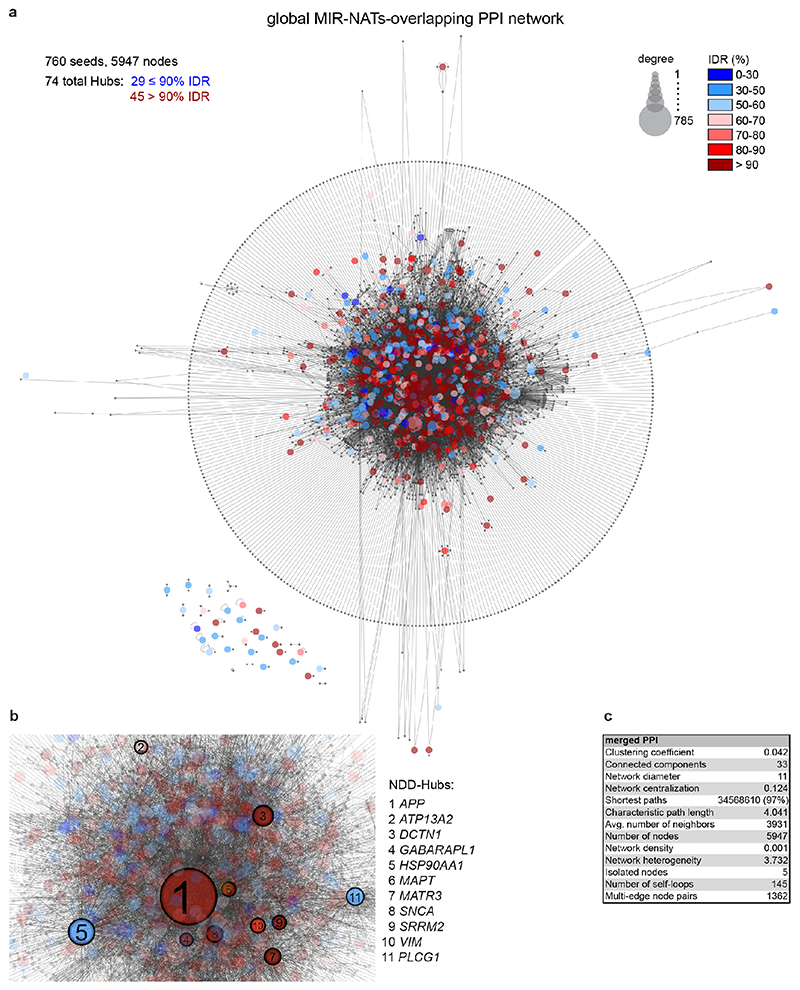
Extended Data Figure 11. Majority of genes targeted by MIR-NATs are enriched for interacting intrinsically disordered proteins (IDPs).
(a) Extended protein-protein interaction (PPI)-network from experimentally validated interaction data from various databases mined by PINOT29, using 760 nonredundant seed proteins participating in S-AS pairs with MIR-NATs. 399 seeds (40.3%) are genes encoding for IDPs with more than 90% IDRs, represented as red-filled circles, are significantly enriched into the network (p=0.0096, 100,000 random simulations in R, Bonferroni, details in Supplementary Table3). Only first-degree interactions are represented. Percentage of sequence predicted to span intrinsically disordered regions (IDRs) by at least 75% of the 9 algorithms from the D2P2 database30 is colour coded from blue (0-30%) to red (>90%) (b), 11 NDD-hub proteins in the above network are presented in this zoom-in view: (APP, ATP13A2, DCTN1, GABARAPL1, HSP90AA1, MAPT, MATR3, PLCG1, SNCA, SRRM2, VIM) (c), Topological properties of extended PPI network, computed by Cytoscape68.