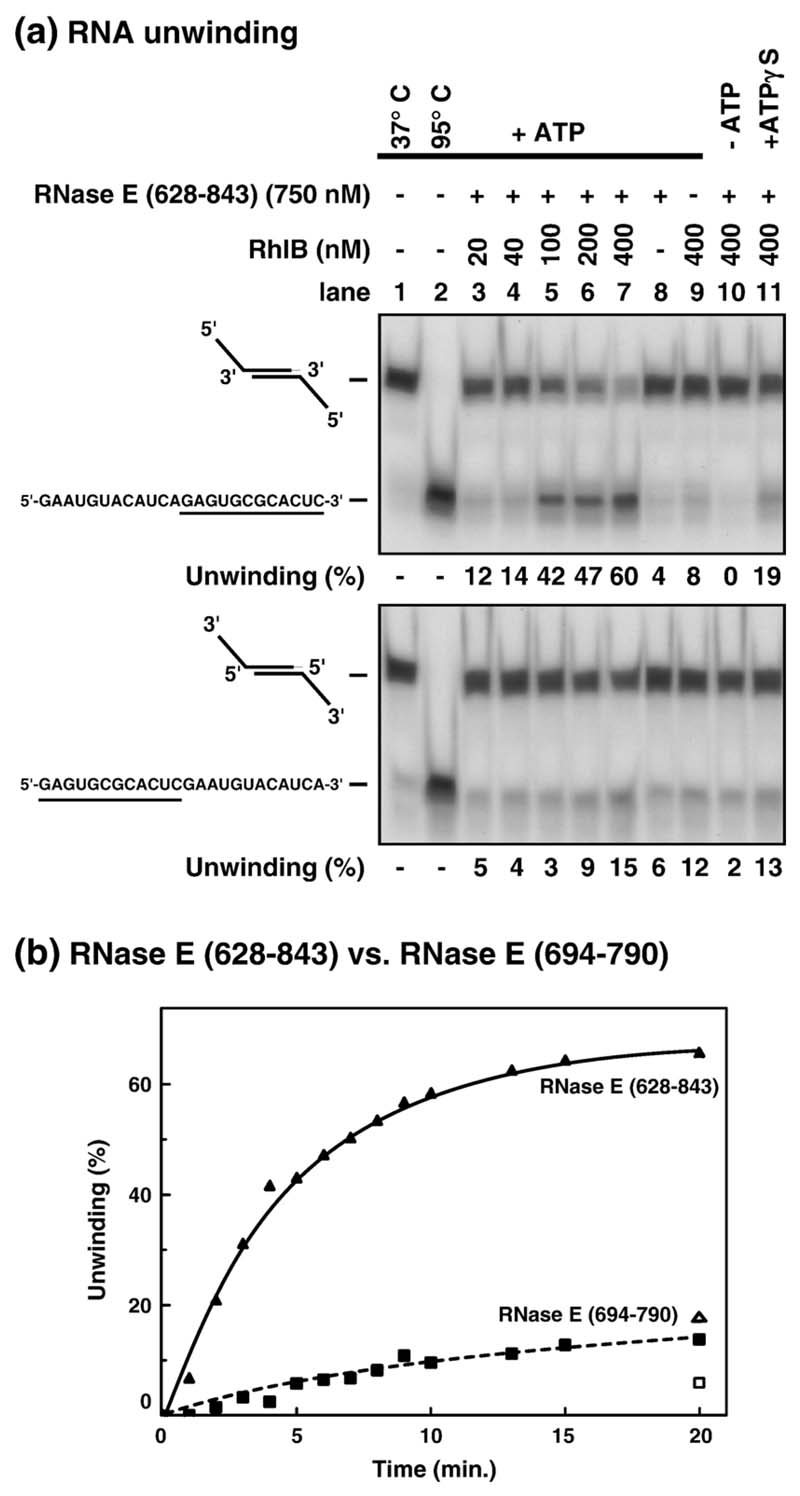
Figure 3. Stimulation of RNA unwinding activity by RNase E (628–843).
(a) RNase E (628-843) and RhlB were incubated as described in Materials and Methods with 5' or 3'-tailed duplex RNA (0.1 nM) for 30 min at 37 °C (upper and lower panels, respectively). Each substrate was formed from a self-annealing synthetic RNA oligonucleotide whose sequence is indicated on the left. The sequence of the single-strand and duplex forming (underlined) region of both substrates is identical. The oligonucleotides were 5'-labelled with γATP. Unwinding was analysed by native PAGE (12%) and autoradiography. The unwinding was quantified using a phosphorimager and is indicated as a percentage under each panel. Lane 1 shows duplex RNA incubated under unwinding conditions but without protein. The percentage of single-stranded RNA in this control (typically 5%) was subtracted as a background in the unwinding reactions. Lanes 2 shows duplex RNAs denatured by heating to 95 °C for 3 min. (b) The stimulation of RhlB unwinding activity by RNase E (694–790) is comparatively less efficient. 400 nM RhlB and 0.1 nM 5'-tailed duplex were incubated with 750 nM RNase E (628–843) (filled triangle) or 660 nM RNase E (694-790) (filled square) at 37 °C with ATP. In the presence of ATPγS, 17% (open triangle) and 4% (open square) of single-stranded substrate is produced after 20 min with RNase E (628–843) and RNase E (694–790), respectively, compared to 66% and 14% with ATP.