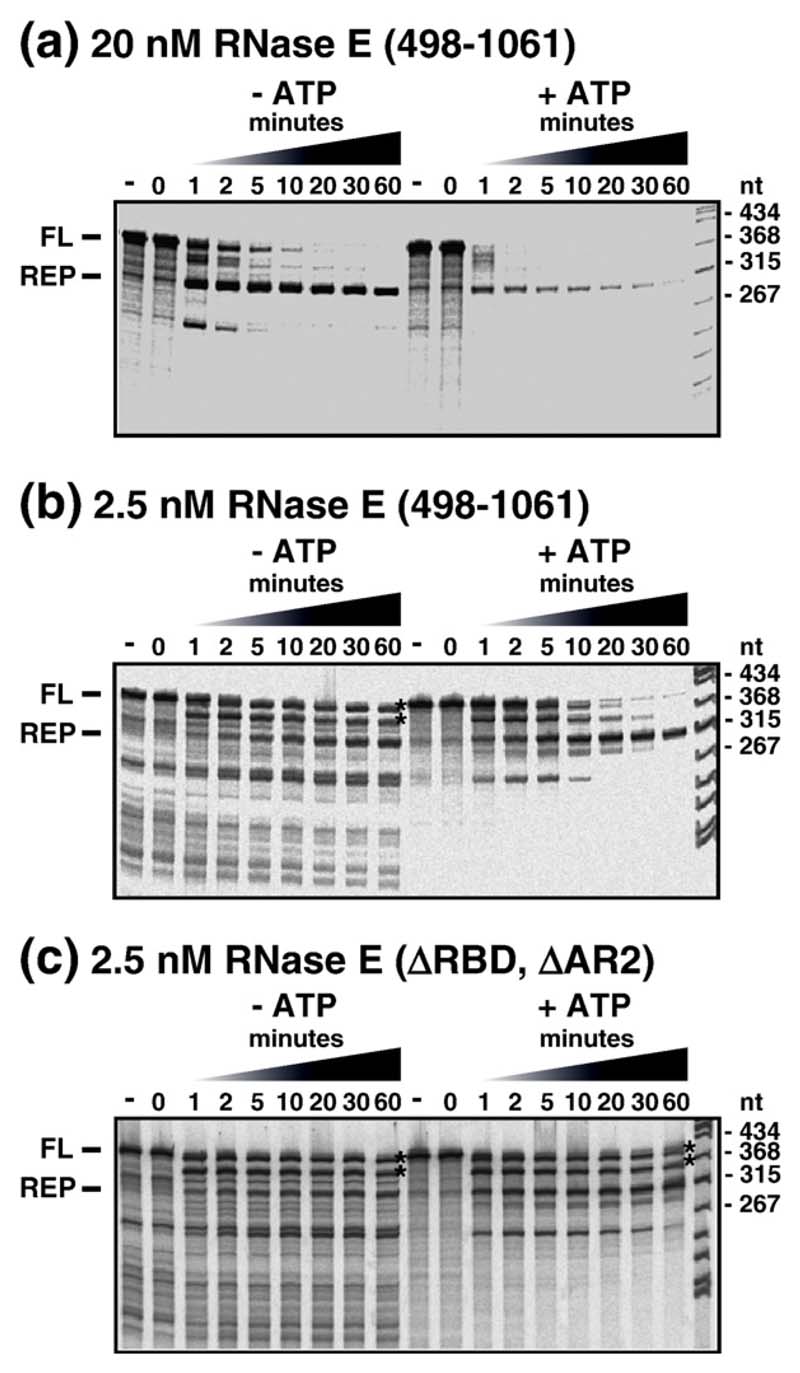
Figure 5.
Helicase-facilitated RNA degradation in the mini-degradosome. Mini-degradosomes were employed in the degradation of an mRNA fragment derived from the intergenic region of the malEF operon containing two repetitive extragenic palindrome (REP) sequences (see the text). The panels show autoradiograms in which the products of digestion in a time course were separated by denaturing gel electrophoreses (6% (w/v) acrylamide, 29:1, 8 M urea). To the left of each panel, FL indicates the full-length substrate and REP, the intermediate in which degradation arrests a few nucleotides 3' of the REP structure. In each panel, two time courses are presented: in the absence of ATP (left) and in the presence of ATP (right). Mini-degradosomes were assembled from RhlB, PNPase and RNase E (498–1061) or its derivatives. (a) Degradation reaction using a mini-degradosome assembled from intact RNase E (498–1061). The concentration of the complex, based on PNPase content, was 20 nM in a reaction containing 20 nM substrate. (b and c) Degradation reactions with 2.5 nM complex and 20 nM substrate in which the conditions had been altered to reduce the activity of PNPase (see the text).