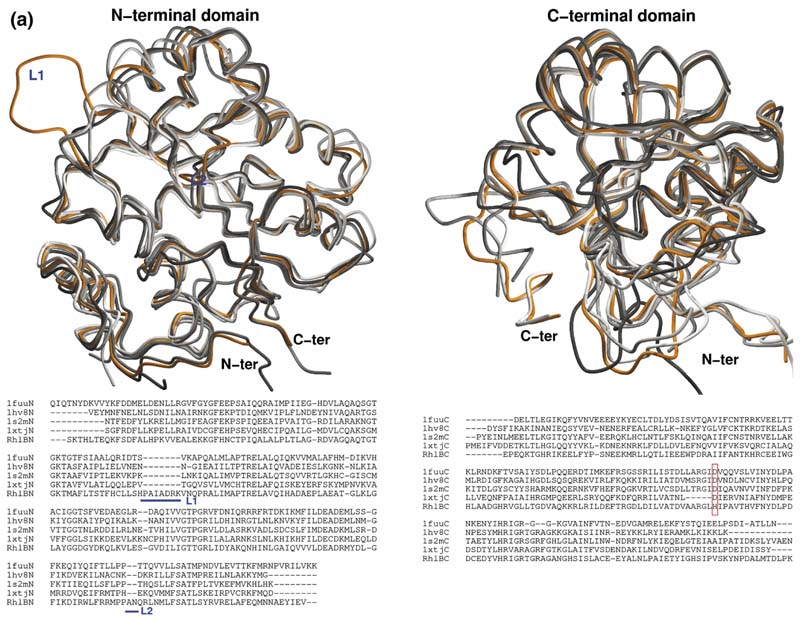
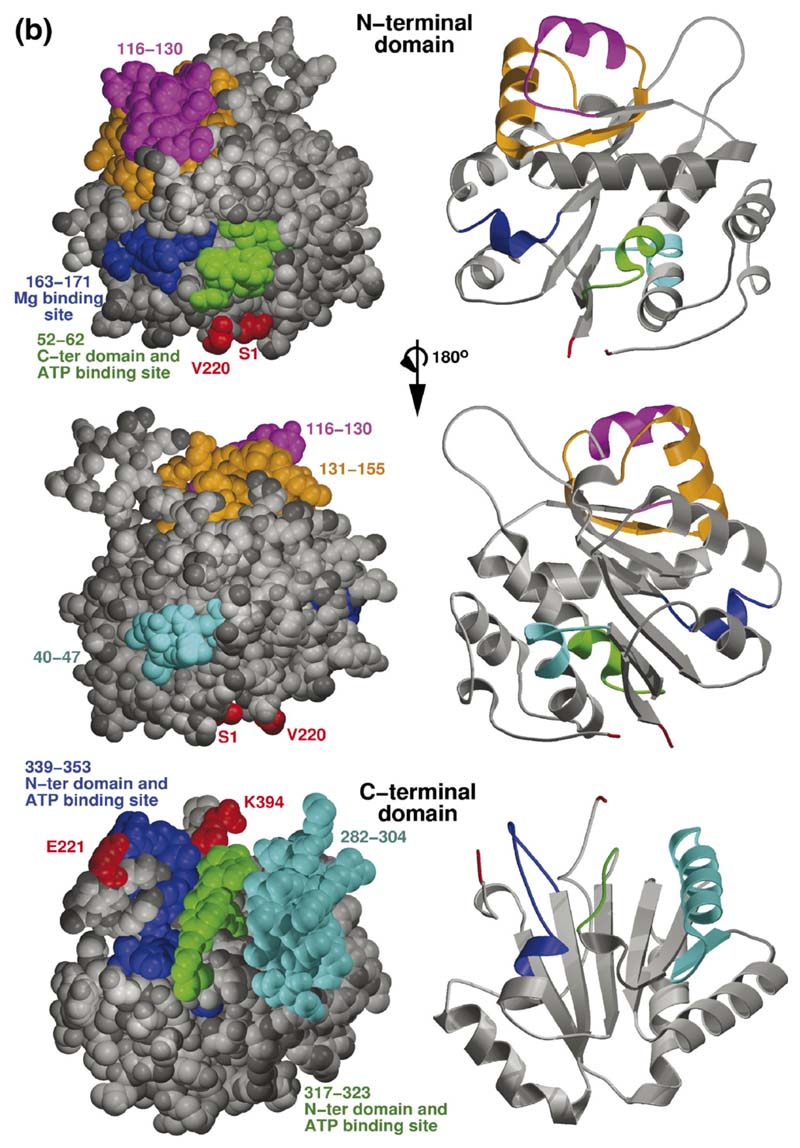
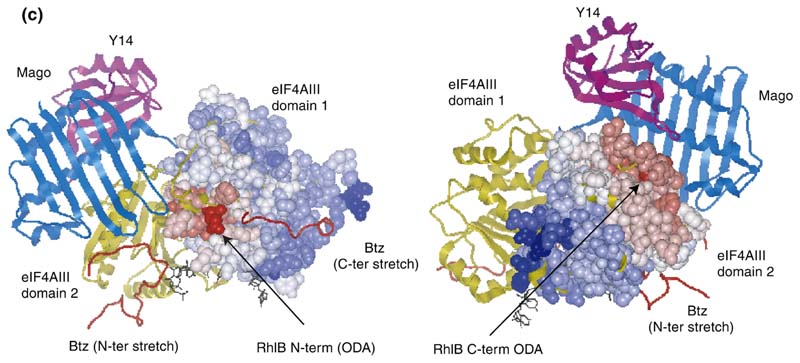
Figure 6. Homology model of RhlB and the predicted interaction site with RNase E.
(a) Overlay of the independent domains with the templates (grey) and predicted models (orange) for the NTD and CTD of RhlB, indicating the differences in the loops. The inset shows the structure-based sequence alignment of the structural homologues, with the inserted loops indicated. The boxed region shows a conserved difference between RhlB and its homologues in motif V (see Supplementary Data, Figure S2 for sequence alignment of RhlB proteins from other Gram-negative bacteria in which the RNase E has a conserved potential helicase binding site). (b) Functional patterns on the surface of RhlB predicted by the program FPSPD.40 Three functional patterns are predicted on the surface of the C-terminal domain and five on the surface of the N-terminal domain. Pattern 52-62 is predicted to be involved in interactions with the C-terminal domain and ATP, and pattern 163-171 is most likely involved in interactions with the magnesium ion. Patterns 40-47,116-130 and 131-155 are probably not involved in domain interactions and may be involved in other interactions. Residues in red correspond to the N and C termini for each domain. (c) Comparison of the optimal docking area (ODA) identified on RhlB with the protein-protein interacting region on eIF4AIII in the exon junction complex (pdb code 2JOS).11 The left panel shows an overlay of the RhlB-NTD with eIF4AIII-domain 1 (RMSD 2.3 Å) and the right panel shows the overlay of RhlB-CTD with eIF4AIII-domain 2 (RMSD 3.3 Å). RhlB residues are shown as CPK spheres here. The ODA identified on both domains of RhlB is shown in red.