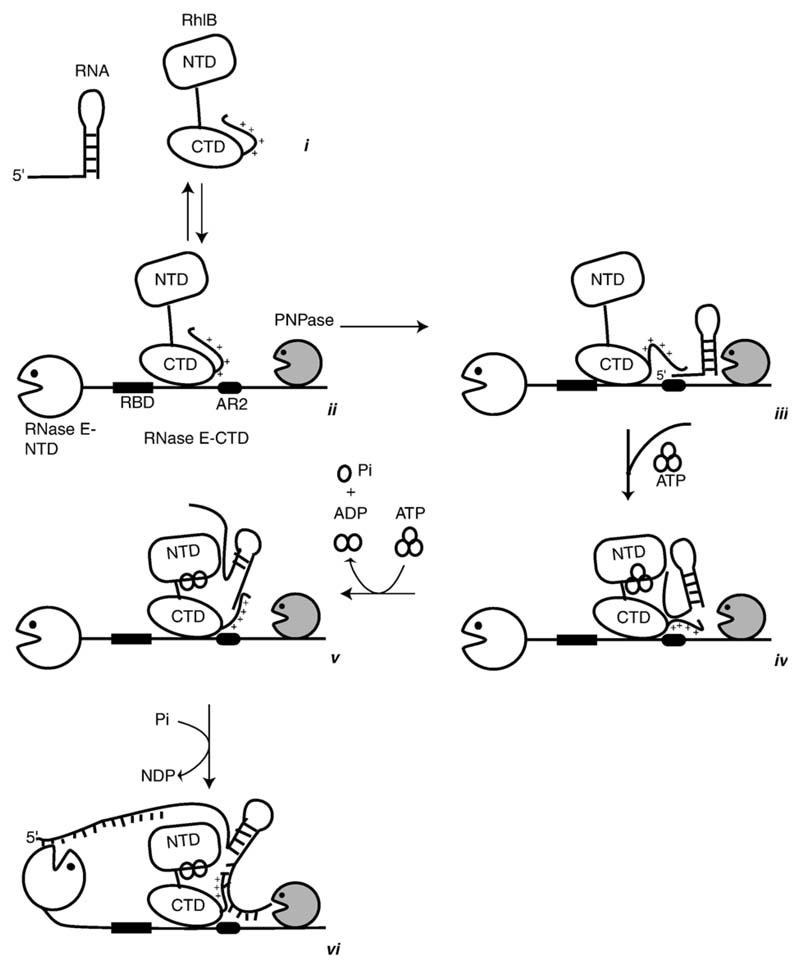
Figure 7.
A model for the sequential steps in the unwinding and translation mechanism of the cooperating helicase-ribonuclease assembly. The helicase is envisaged as a two-domain structure with a flexible inter-domain linker and positively charged C-terminal tail. The isolated helicase interacts with RNA (i), but may interact only weakly while engaged to RNase E (ii). In the cartoon, PNPase is also engaged with RNase E (ii). RNA may bind to RNase E at RBD or AR2 (as indicated here by interaction with the stem-loop RNA structure, (iii) or both. Cycles of ATP binding and hydrolysis may expose the surfaces of the RecA-like domains of RhlB to interact with the RNA and stabilise its interaction during unwinding and 5' to 3' translocation (iv) and (v). The exposed terminal regions of the RNA are subject to phosphorolytic cleavage by the adjacent PNPase or hydrolytic attack internally by the catalytic domain of RNase E (vi). See the text for further narrative.