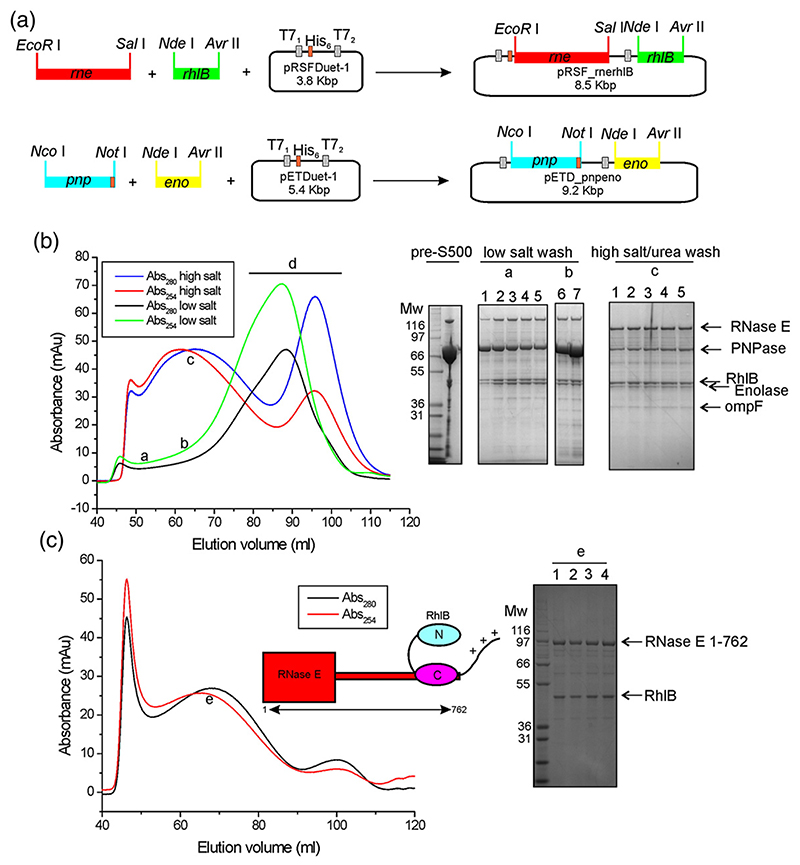
Fig. 2. Schematic of the cloning strategy and purification results of recombinant degradosome assemblies.
(a) DNAs encoding for RNase E and RhlB were inserted into the vector pRSFDuet-1; C-terminal histidine-tagged PNPase, together with enolase, were inserted into a pETDuet-1 vector. (b) The elution profile from a preparative S500 size-exclusion column, comparing samples from low-salt or high-salt/urea washes at the Ni2+-affinity matrix step. SDS-PAGE shows an example of a concentrated pre-S500 fraction and of fractions collected at certain elution volumes for the different S500 runs: (a and b) low-salt wash, 4–12% polyacrylamide bis-Tris gel (lanes 1–5, 50–54 ml; lanes 6–7, 75 and 80 ml); (c) high-salt/urea wash, 10% polyacrylamide bis-Tris (lanes 1–5,65–77 ml). In the portions of the chromatogram labelled “a” and “b” excess PNPase is clearly visible, and the portion “d” contains mainly free PNPase. The faint band migrating below enolase is the outer membrane protein OmpF. Molecular weight markers are indicated in kilodaltons. (c) The S500 elution profile of the C-terminally truncated RNase E 1–762/RhlB subassembly previously subjected to a wash step on a Ni2+-affinity column with high-salt/urea buffer. The sharp peak eluting at ~45 ml contains protein aggregates. The broad peak labelled “e” with a low A254/280 nm ratio was analyzed by SDS-PAGE (lanes 1–4 fractions eluting at 68–71 ml) and was found to contain the degradosome subassembly (cartoon insert).