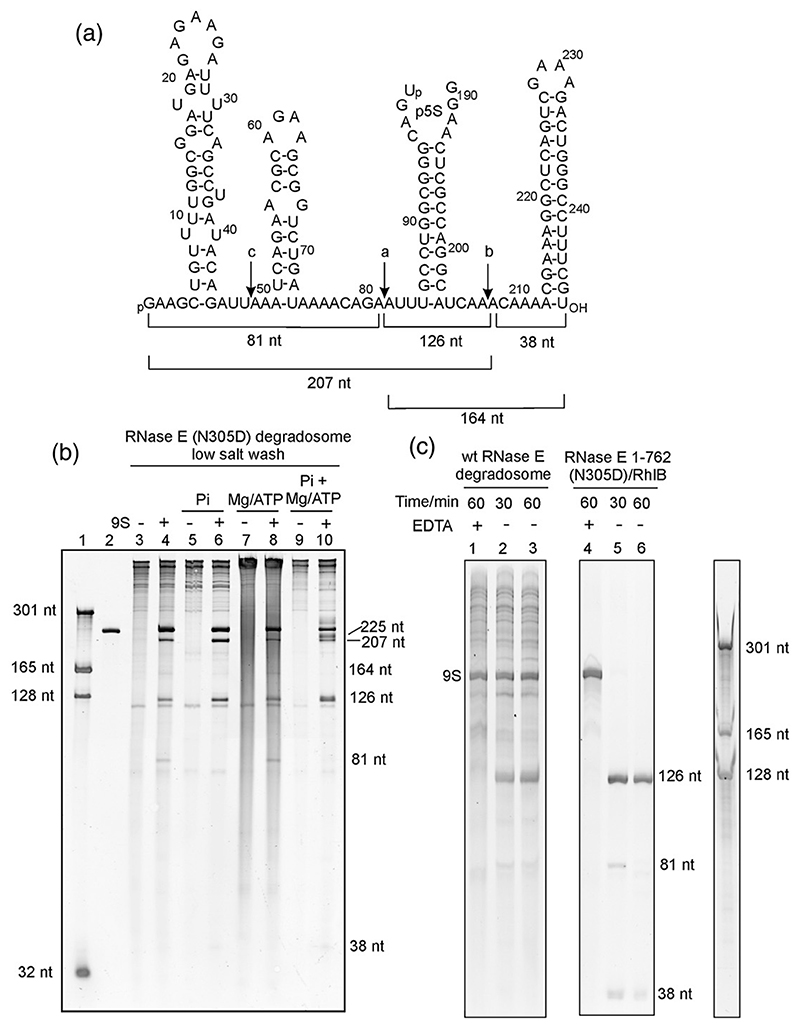
Fig. 4. Processing of 9S RNA.
(a) Sequence and predicted secondary structural elements of 9S RNA. The in-vitro-transcribed 9S RNA used in this work has a G at positions +1 and + 2. The locations of the two major cleavage sites (indicated by the letters a and b in the schematic) and the minor cleavage site (c in the schematic) are indicated with downward arrows. (b and c) Denaturing PAGE gels stained with SYBR Gold reveal the processed products of 9S RNA by the recombinant degradosome and RNase E(1–762)/RhlB subassembly. Lane 1 in (b) represents RNA size markers, and lane 2 represents in-vitro-transcribed 9S RNA. 9S RNA processing was tested in the absence (lanes 3–4) and in the presence (lanes 5–10) of additives. The band at 126 nt corresponds to p5S, a precursor of the mature 120-nt 5S rRNA. In (c), the addition of EDTA is seen to inhibit 9S RNA processing by wild-type (wt) RNase E in the recombinant degradosome assembly (lane 1) and in the RNase E(1–762) RhlB subassembly (lane 4). In the absence of EDTA, processing occurs (lanes 2, 3, 5, and 6). The 9S secondary structure schematic was adopted from Cormack and Mackie.41