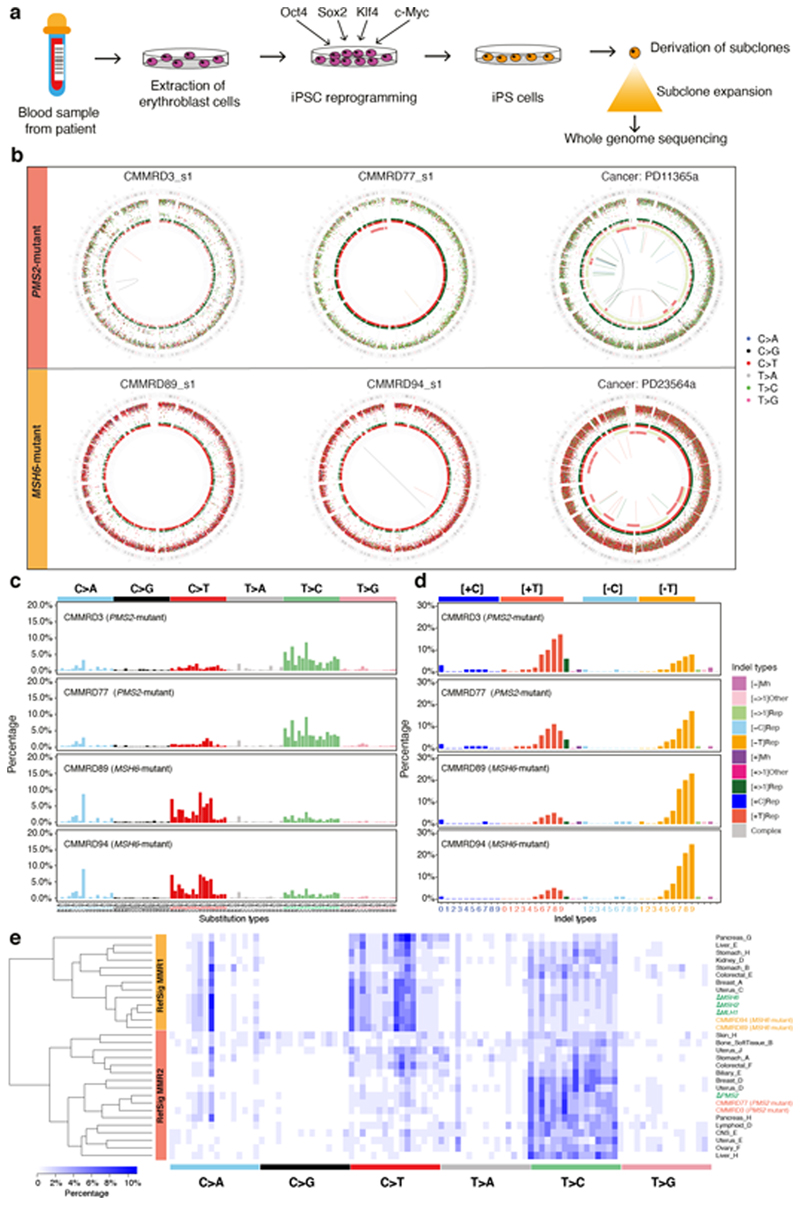
Figure 4. Gene-specific features of signatures of mismatch repair (MMR) deficiency are recapitulated in other model systems.
(a) Experimental workflow including generation of hiPSCs from patients with Constitutional Mismatch Repair Deficiency (CMMRD), subcloning of hiPSCs and whole-genome sequencing. (b) Genome plots of MMR knockouts demonstrate consistent gene-specificity regardless of model system, e.g., cancer (in vivo) and CMMRD patient-derived hiPSCs (in vitro). Top: whole genome plots of two iPSC subclones from two PMS2 mutated CMMRD patients and a breast tumor with PMS2 deficiency. Bottom: genome plots of two iPSC subclones derived from two MSH6 mutant CMMRD patients and a breast tumor with MSH2/MSH6 deficiency. Genome plots show somatic mutations including substitutions (outermost, dots represent six mutation types: C>A, blue; C>G, black; C>T, red; T>A, grey; T>C, green; T>G, pink), indels (the second outer circle, colour bars represent five types of indels: complex, grey; insertion, green; deletion other, red; repeat-mediated deletion, light red; microhomology-mediated deletion, dark red) and rearrangements (innermost, lines representing different types of rearrangements: tandem duplications, green; deletions, orange; inversions, blue; translocations, grey). (c) 96-channel substitution profiles. (d) 45-channel indel profiles. (e) Hierarchical clustering of cancer-derived tissue-specific MMR signature and MMR knockout signatures. 96-bar plots of ΔPMS2-related tissue-specific signatures can be viewed here:
https://signal.mutationalsignatures.com/explore/cancer/consensusSubstitutionSignatures/6