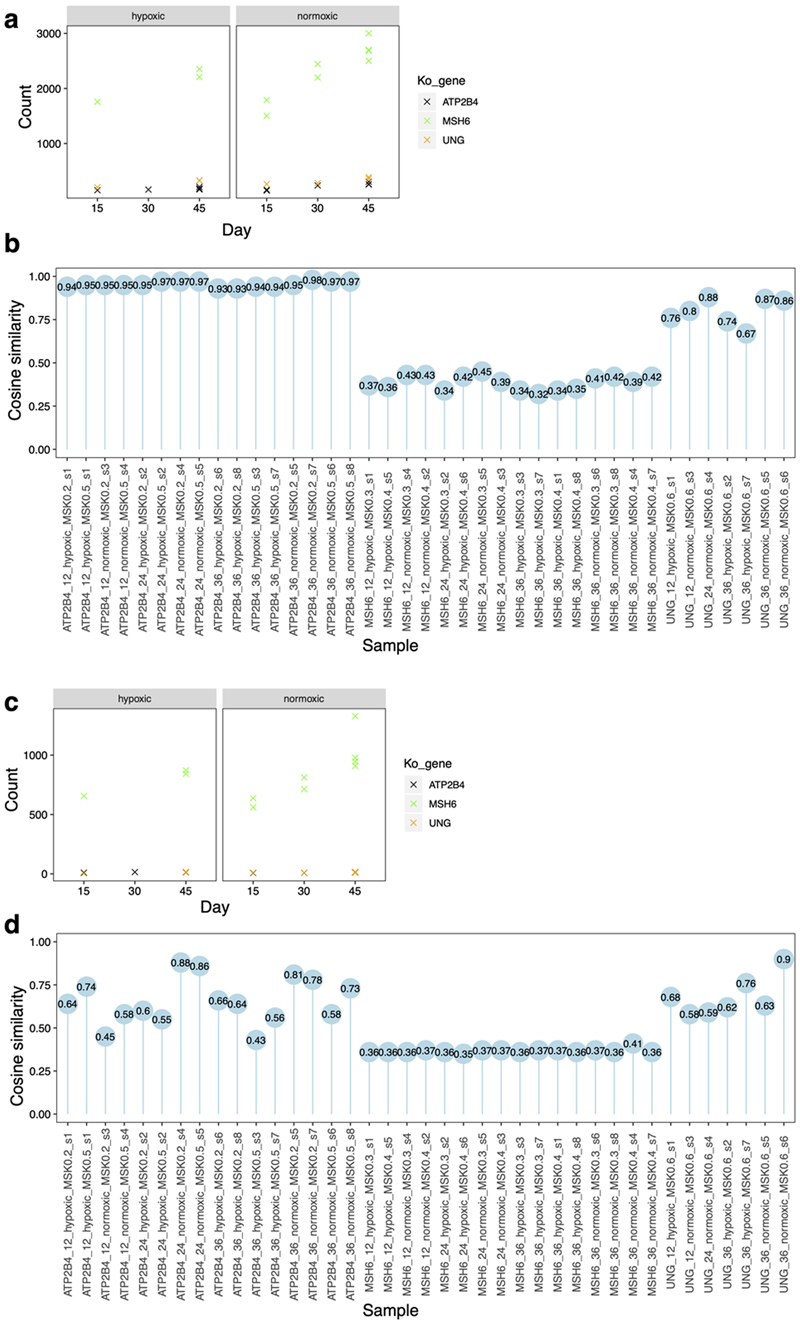
Extended Data Fig. 1. Results of pilot study.
Three genes were selected for knockout (&#Δ): MSH6, UNG and ATP2B4 (negative control). Two genotypes per gene were obtained and grown in culture to gauge reproducibility of signatures between different genotypes of a gene-knockout. These lines were cultured under normoxic (20%) and hypoxic (3%) states, for defined culture times of ~15, 30 or 45 days. Two single-cell subclones were derived for whole genome sequencing for each parental line (equivalent to four subclones per gene edit). One of the UNG genotypes appeared to be heterozygous, which was excluded in downstream analysis. (a) Substitution burden for knockouts of ATP2B4, UNG and MSH6 under hypoxic and normoxic conditions as well as different culturing time. (b) The cosine similarities between the mutational profile of each subclone and background signature of culture. (c) Indel burden for knockouts of ATP2B4, UNG and MSH6 under hypoxic and normoxic conditions as well as different culturing time. (d) The cosine similarities between the mutational profile of each subclone with background signature of culture. Overall, the differences between normoxic and hypoxic conditions were not marked, although normoxic conditions produced slightly more mutations. Time in culture made only a marginal, non-linear difference to burden of mutagenesis. Given the results of the pilot, weighing up the costs and risks associated with prolonged culture time (risk of infection, risk of selection, marked increase in cost of experimental reagents) with the minimal return in terms of mutation number, and also intending to minimize transitions between hypoxic to normoxic conditions while handling cell cultures, we opted to proceed with the full-scale study under normoxic conditions and for 15 days for the rest of study.