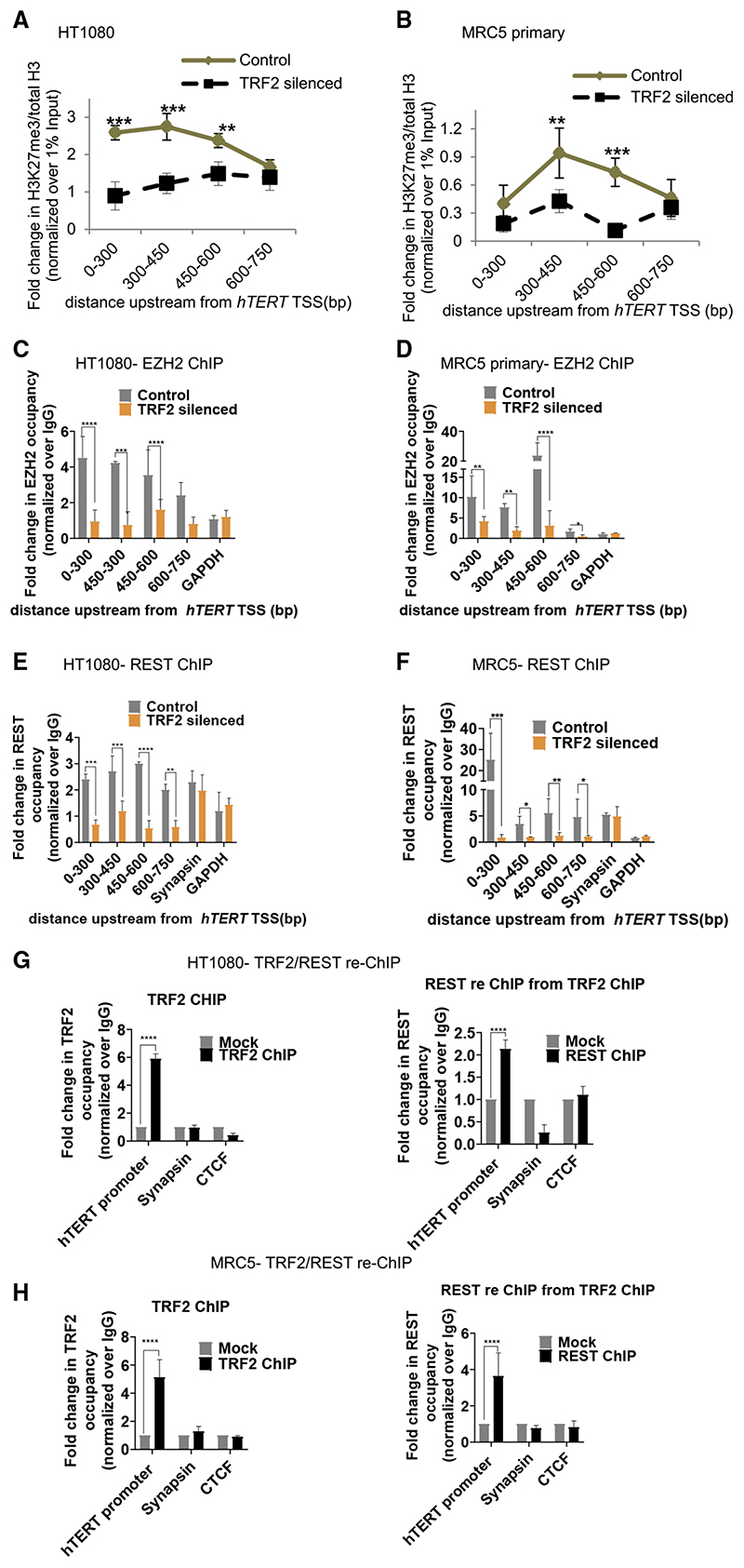
Figure 2. TRF2 recruits the polycomb repressor complex (PRC2) at the hTERT promoter.
(A and B) Effect of TRF2 silencing on H3K27me3 occupancy (ChIP-qPCR) spanning 0–750 bpofthe hTERT promoterHT1080
(A) and MRC5 (B) cells. Fold change represented as H3K27me3 ChIP over total H3 ChIP, normalized to 1% input in respective cases (see STAR Methods for detail).
(C and D) EZH2 occupancy on the hTERT promoter (spanning 0–750 bp) on silencing TRF2 in HT1080 (C) and MRC5 cells (D). Scrambled siRNA-treated cells as control.
(E and F) REST occupancy on the hTERT promoter on silencing TRF2 in HT1080 (E) and MRC5 (F) cells. Synapsin promoter reported for REST binding was used as control forTRF2-inde-pendent REST occupancy. Scrambled siRNA-treated cells as control.
(G and H) TRF2 ChIP followed by REST re-ChIP: TRF2 ChIP (left panel) and REST re-ChIP (right panel) in HT1080 (G) and MRC5 (H) cells at the hTERT core promoter (+38 to −237 bp). Sya-napsin, where REST binding is independent of TRF2 used as control for TRF2-REST co-binding in TRF2/REST-re-ChIP. GAPDH across replicates was not detectable following reChIP therefore CTCF used as negative control for reChIP experiments. All error bars represent ± SDs from mean values; p values calculated by paired/unpaired t test, for (A)–(F) two-way ANOVA was used (*p < 0.05, **p < 0.01, ***p < 0.005, ****p < 0.0001).