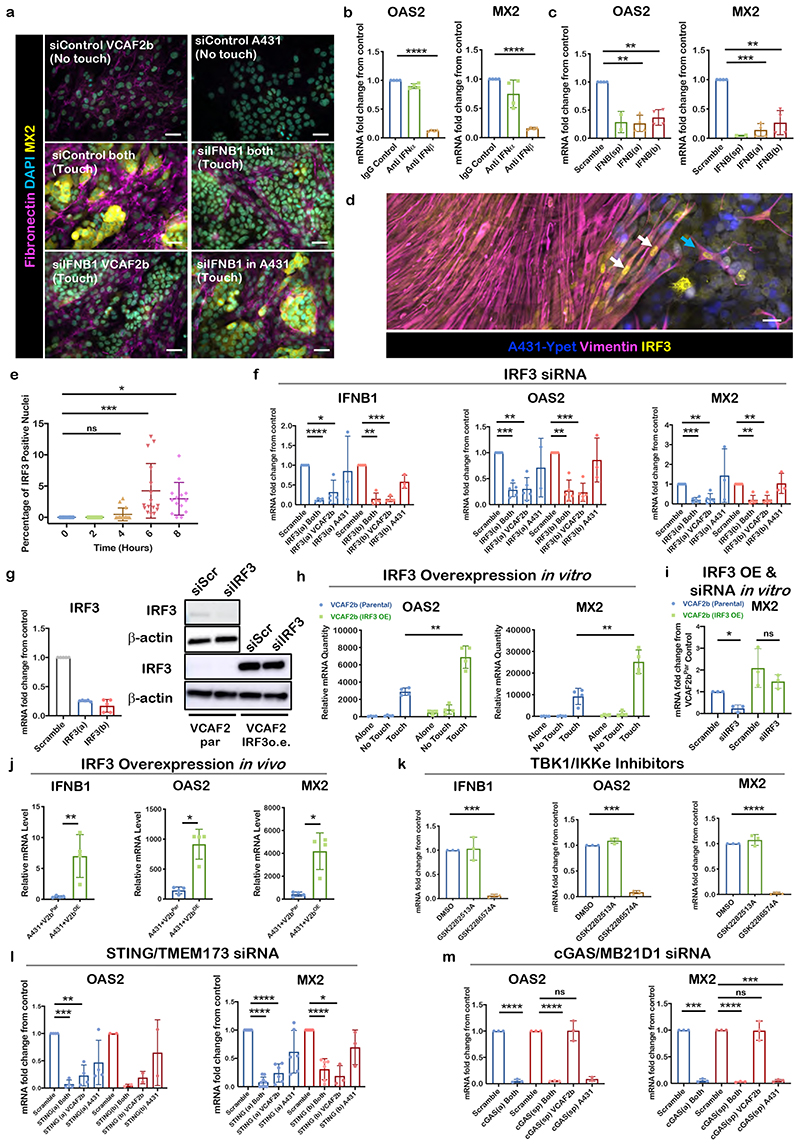
Figure 2. Cytoplasmic DNA mechanisms converge on IRF3 to drive ISG expression following cancer cell-CAF contact.
A) Micrographs of IF staining of different culture conditions after cell-type specific siRNA knockdown (KD) of IFNB1. Fibronectin (magenta), MX2 (yellow), DAPI (cyan). Shown representative images from 2 biological replicates. Scale bar is 50 μm. B) qRT-PCR of ISGs MX2 and OAS2 after co-culture with IFN type-1 specific blocking antibodies. mRNA levels shown were normalized to IgG control. Each dot is a technical replicate from two biological replicates. One sample t test, OAS2 anti-IFNβ vs IgG control Theoretical mean 1, Actual mean 0.1288, Discrepancy -0.8712, 95% CI -0.8884 to -0.8539, t=160.7, df=3, p<0.0001. MX2 anti-IFNβ vs IgG control Theoretical mean 1, Actual mean 0.1606, Discrepancy - 0.8394, 95% CI -0.8652 to -0.8136, t=103.6, df=3 p<0.0001. C) qRT-PCR of ISGs MX2 and OAS2 after siRNA KD of IFNB1 of both cell types using a pool of two separate siRNA sequences, and then each siRNA individually (a, b). Each dot is a biological replicate (n=4). mRNA levels shown were normalized to siRNA scramble control. One sample t test, OAS2 siIFNB1(a) Theoretical mean 1, Actual mean 0.2652, Discrepancy -0.7348, 95% CI -0.9650 to -0.5047, t=10.16, df=3, p=0.0020. siIFNB1(b) Theoretical mean 1, Actual mean 0.3697, Discrepancy -0.6303, 95% CI -0.8516 to -0.4090, t=9.063, df=3, p=0.0028. MX2 siIFNB1(a) Theoretical mean 1, Actual mean 0.1432, Discrepancy -0.8568, 95% CI -1.030 to -0.6838, t=15.76, df=3, p=0.0006. siIFNB1(b) Theoretical mean 1, Actual mean 0.2674, Discrepancy - 0.7326, 95% CI -1.060 to -0.4049, t=7.113, df=3, p=0.0057. D) Micrographs of IF stainings from A431/VCAF2b co-culture with defined borders (silicone inserts used) showing nuclear translocation of IRF3 in VCAF2b upon contact with A431. White arrows highlight nuclear IRF3 signal in VCAF2b in the boundary. Phalloidin (grey), Keratin (magenta), IRF3 (yellow), DAPI (cyan), Shown representative images from two biological replicates. Scale bar is 50μm. E) Quantification of micrographs from A431-VCAF2b co-culture stained with IRF3, over a time course. Hours indicating time passed after addition of A431 cells to VCAF2b. Depicted is the percentage of IRF3 positive nuclei of VCAF2b cells. Each dot indicates a 20x field of view from four different technical replicates. One-way ANOVA with Sidak’s multiple comparisons test. ANOVA, F=8.681, R square=0.3553 df=67, p<0.0001. 0hvs4h Mean diff - 0.4756, 95% CI -2.966 to 2.015, p=0.9538, 0hvs6h Mean diff -4.244,95% CI -6.703 to - 1.785, p=0.0002, 0hvs8h Mean diff -2.961,95% CI -5.487 to -0.4354, p=0.0164. F) qRT-PCR of, IFNB1, OAS2 and MX2 in direct A431/VCAF2b co-cultures after cell-type specific siRNA KD of IRF3 using two different sequences. Each dot is a biological replicate. mRNA levels shown were normalized to siRNA scramble control. One sample t test, IFNB1 siIRF3(a) Both Theoretical mean 1, Actual mean 0.1207, Discrepancy -0.8793, 95% CI -0.9368 to -0.8218, t=48.65, df=3, p<0.0001. siIRF3(a) VCAF2b Theoretical mean 1, Actual mean 0.3201, Discrepancy -0.6799, 95% CI -1.159 to -0.2011, t=4.519, df=3, p=0.0203. siIRF3(b) Both : Theoretical mean 1, Actual mean 0.1486, Discrepancy -0.8514, 95% CI -1.086 to -0.6171, t=11.56, df=3, p=0.0014. siIRF3(b) VCAF2b Theoretical mean 1, Actual mean 0.1443, Discrepancy -0.8557, 95% CI -0.9640 to -0.7475, t=25.16, df=3, p<0.001. OAS2 siIRF3(a) Both, Theoretical mean 1, Actual mean 0.2840, Discrepancy -0.7160, 95% CI -0.8788 to - 0.5532, t=12.21, df=4, p=0.0003. siIRF3(a) VCAF2b, Theoretical mean 1, Actual mean 0.3033, Discrepancy -0.6967, 95% CI -0.9663 to -0.4271, t=7.176, df=4, p=0.0020. siIRF3(b) Both, Theoretical mean 1, Actual mean 0.2698, Discrepancy -0.7302, 95% CI -0.9827 to - 0.4777, t=8.028, df=4, p=0.0013, siIRF3(b) VCAF2b p=0.0006, siIRF3(b) A431 Theoretical mean 1, Actual mean 0.3201, discrepancy -0.6799, 95% CI -1.159 to -0.2011, t=4.519, df=3, p=0.6231. MX2 siIRF3(a) Both p<0.0001, siIRF3(a) VCAF2b p=0.0028, siIRF3(a) A431 p=0.6327. siIRF3(b) Both p=0.0014, siIRF3(b) VCAF2b, Theoretical mean 1, Actual mean 0.2370, Discrepancy -0.7630, 95% CI -0.9808 to -0.5452, t=9.728, df=4. p=0.0012. G) IRF3 siRNA Knockdown efficiency shown through both qRT-PCR (left) and Western blot (right). qRT-PCR of IRF3 expression in VCAF2b cells after treatment with either scrambled control or two different IRF3 siRNA (a - red, b - blue). Each dot is a biological replicate (n=4) and mRNA levels shown were normalized to siRNA scramble control. Top panel of western blot shows IRF3 expression in VCAF2b. Bottom panels show IRF3 expression in VCAF2b parental (VCAF2b par) and lentiviral transfected VCAF2b with siRNA resistant IRF3 (VACF2b o.e.) (bottom). To avoid saturating the signal for the over-expressed IRF3, the lower panel is a shorter exposure and hence the endogenous protein is not easily visible. Representative blot of two biological replicates. IRF3 (50-55kDa) and loading control Antibeta actin (43kDa). H) qRT-PCR of OAS2 and MX2 in direct, indirect co-cultures and cells on their own using A431 and VCAF2 parental (VCAF2b par) and VCAF2b transfected with IRF3 construct (VCAF2b IRF3 OE). mRNA levels were normalized against two housekeeping genes. Each dot represents a biological replicate (n=4). Unpaired t test, Touch parental vs Touch IRF3 OE OAS2, Mean difference -680.8, SE of diff 680.8, T=5.87, df=6, p=0.0010. MX2, Mean difference -16016, SE of diff 3308, T=4.842, df=6, p=0.0028 I) qRT-PCR of MX2 of direct co-cultures of A431 with VCAF2b par and VCAF2b OE after transfection with IRF3 siRNA. Each dot is a biological replicate (n=3). mRNA levels shown were normalized to siRNA scramble control. One sample t test, VCAF2b par siIRF3, Theoretical mean 1, Actual mean 0.2351, Discrepancy -0.7649, 95% CI -1.161 to -0.3686, t=8.305, df=2, p=0.0142. Unpaired t test VCAF2b OE Scramble vs siIRF3, mean difference - 0.6156, 95% CI -2.128 to 0.8966, t=1.13, df=4. p=0.3215 J) qRT-PCR of IFNB1, OAS2 and MX2 from subcutaneous tumours four days after injection of A431-VCA2b par or A431-VCAF2b OE in the flank of Balb/c nude mice. Each dot represents a sample from an individual tumour (n=4). mRNA levels were normalized against two housekeeping genes. Unpaired t test IFNB1, Mean difference 6.578, 95%CI of diff. 2.33 to 10.83, T=3.789, df=6, p=0.0091. Mann Whitney test, OAS2, diff btw medians 877.7, p=0.0286 MX2, diff btw medians 4430 p=0.0286 K) qRT-PCR of IFNB1, OAS2 and MX2 in direct co-cultures after treatment with DMSO, structural control agent (GSK2282513A) or TBK/IKKε inhibitor (GSK2286574A). Each dot is a biological replicate (n=3). mRNA levels shown were normalized to DMSO control. One sample t test for GSK2286574A IFNB1 Theoretical mean 1, Actual mean 0.06098, Discrepancy -0.939, 95% CI -1.009 to -0.8688, t=57.53, df=2, p=0.0003, OAS2 Theoretical mean 1, Actual mean 0.08724, Discrepancy -0.9128, 95% CI - 0.9914 to -0.8341, t=49.93, df=2, p=0.0004. MX2 Theoretical mean 1, Actual mean 0.02398, Discrepancy -0.976, 95% CI -1.024 to -0.9282, t=87.82, df=2, p=0.0001. L) qRT-PCR of OAS2 and MX2 after siRNA KD of two different TMEM173/STING sequences in both cell types and cell-type specific. Each dot is a biological replicate. mRNA levels shown were normalized to siRNA scramble control. One sample t test OAS2 STING(a) Both, Theoretical mean 1, Actual mean 0.07346, Discrepancy -0.9265, 95% CI -1.046 to -0.8073, t=24.72, df=3, p<0.0001, STING(a) VCAF2b, Theoretical mean 1, Actual mean 0.2311, Discrepancy - 0.7689, 95% CI -1.074 to -0.4636, t=8.014, df=3, p=0.0041. STING(a) A431, Theoretical mean 1, Actual mean 0.4696, Discrepancy -0.5304, 95% CI - -1.177 to 0.1164, t=2.61, df=3 p=0.0797. MX2 STING(a) Both, Theoretical mean 1, Actual mean 0.07983, Discrepancy - 0.9202, 95% CI -0.9891 to -0.8513, t=30.80, df=8, p<0.0001. STING(a) VCAF2b, Theoretical mean 1, Actual mean 0.2416, Discrepancy -0.7584, 95% CI -0.9282 to -0.5886, t=11.48, df=5, p<0.0001. STING(a) A431, Theoretical mean 1, Actual mean 0.6166, Discrepancy - 0.3834, 95% CI -0.7782 to 0.01149, t=2.496, df=5, p=0.0548. STING(b) Both, Theoretical mean 1, Actual mean 0.3092, Discrepancy -0.6908, 95% CI -0.8864 to -0.4953, t=9.083, df=5, p=0.0003, STING(b) VCAF2b, Theoretical mean 1, Actual mean 0.1898, Discrepancy - 0.8102, 95% CI -1.262 to -0.3582, t=7.712, df=2, p=0.0164. STING(b) A431, Theoretical mean 1, Actual mean 0.6952, Discrepancy -0.3048, 95% CI - -1.066 to 0.4562, t=1.723, df=5, p=0.2270. M) qRT-PCR of OAS2 and MX2 in direct A431/VCAF2b co-cultures after cell-type specific siRNA KD of cGAS/MB21D1. Each dot is a biological replicate (n=3). mRNA levels shown were normalized to siRNA scramble control. OAS2 cGAS(a) Both, Theoretical mean 1, Actual mean 0.05284, Discrepancy -0.9472, 95% CI -1.021 to -0.8731, t=55.06, df=2, p=0.0003. cGAS(sp) Both, Theoretical mean 1, Actual mean 0.05240, Discrepancy -0.9476, 95% CI -0.9661 to -0.9291, t=220.7, df=2, p<0.0001. cGAS(sp) VCAF2b, Theoretical mean 1, Actual mean 1.007, Discrepancy 0.006660, 95% CI -0.4635 to 0.4769, t=0.06095, df=2, p=0.9569. MX2 cGAS(a) Both, Theoretical mean 1, Actual mean 0.05284, Discrepancy -0.9472, 95% CI -1.021 to -0.8731, t=55.06, df=2, p=0.0003. cGAS(sp) Both Theoretical mean 1, Actual mean 0.02928, Discrepancy -0.9707, 95% CI - 0.9944 to -0.9470, t=176.2, df=2, p<0.0001. cGAS(sp) VCAF2b, Theoretical mean 1, Actual mean 0.9911, Discrepancy -0.008935, 95% CI -0.4588 to 0.4409, t=0.08546, df=2, p=0.9397. cGAS(sp) A431, Theoretical mean 1, Actual mean 0.05162, Discrepancy - 0.9484, 95% CI -1.006 to -0.8906, t=70.57, df=2, p=0.0002. For all graphs, represented is mean with SD. Statistics: ns not significant, * p<0.05, ** p<0.005, *** p<0.001, **** p<0.0001.