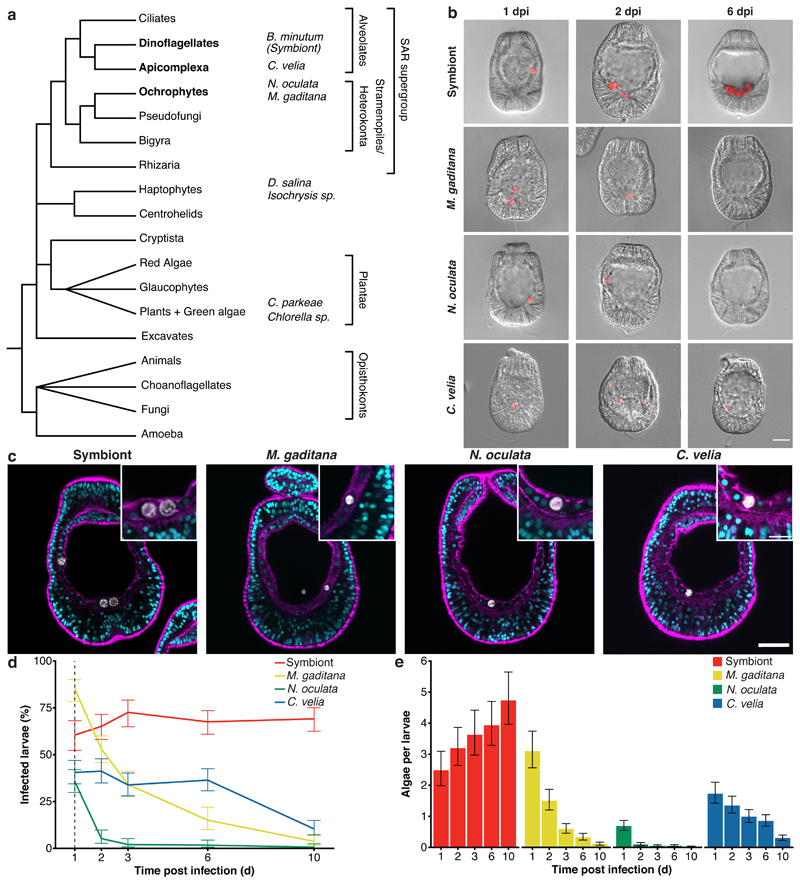
Fig. 1. Aiptasia larvae as a comparative system to dissect symbiont maintenance.
a Phylogenetic tree adapted from 96. The four microalgae used for the comparative analysis belong to the SAR supergroup: B. minutum strain SSB01 (symbiont) belongs to the phylum Dinoflagellata, C. velia belongs to the phylum Apicomplexa, both within Alveolata; while both M. gaditana and N. oculata are members of Ochrophyta. The additional microalgae screened belong to two different phyla: D. salina and Isochrysis sp. - Haptophyta and C. parkeae and Chlorella sp. - Chlorophyta (images in Extended Data Fig. 1). b Aiptasia larvae 1-, 2-, and 6-day(s) post infection (dpi) with symbiont, M. gaditana, N. oculata, and C. velia, as indicated. Larvae were infected at 4-6 days post fertilization (dpf) for 24 hours and were washed into fresh filtered artificial seawater (FASW). Images are merges of DIC (gray) and autofluorescence of microalgae photosynthetic pigments (red), scale bar represents 25 μm (exemplary images from data in d and e). c All microalgae, symbiont, M. gaditana, N. oculata, and C. velia., were taken up in the endodermal tissue. Autofluorescence of microalgal photosynthetic pigment (white), Hoechst-stained nuclei (cyan), phalloidin-stained F-actin (magenta). Scale bar for whole larva represents 25 μm and scale bar for close up represents 10 μm (n=5 with at least 5 larvae imaged each time). d Percentage of Aiptasia larvae infected after exposure to 1.0 x 105 cells ml-1 symbiont, M. gaditana, N. oculata, or C. velia for 24 hours followed by a washout into fresh FASW (indicated by the arrow and dotted line). Error bars indicate mean ± 95 % CI of 5 independent replicates. e Average number of microalgae/larva after exposure to 1.0 x 105 cells ml-1 of symbiont, M. gaditana, N. oculata, or C. velia for 24 hours after which larvae were washed into fresh FASW. Error bars are mean ± 95 % CI of 5 independent replicates with around 50 larvae each.