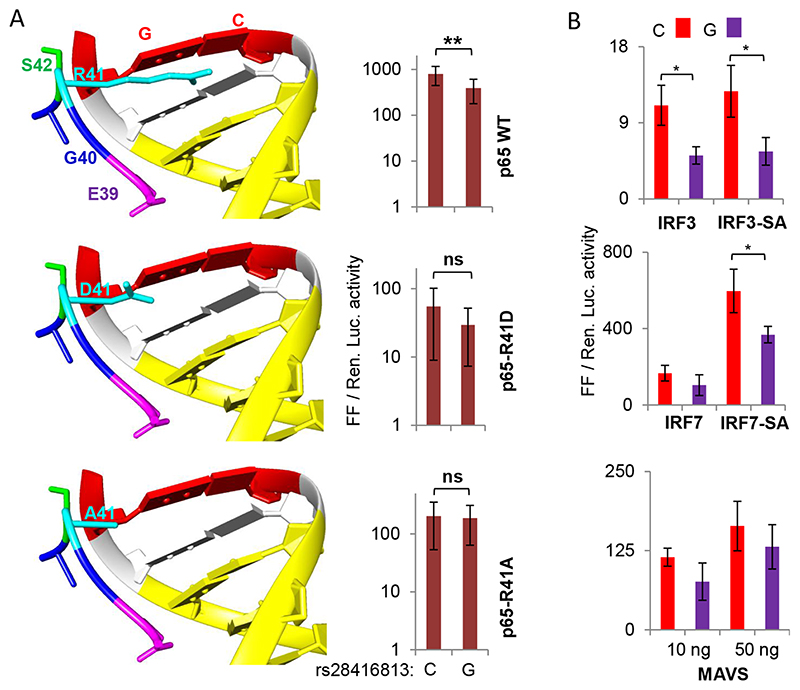
Fig. 2.
A) (Left) R41 of NF-κB interacts with SNP rs28416813. The models show cocrystal structures of NF-κB p50-p65 heterodimers and their cognate DNA together (PDB ID2O61 visualized in UCSF CHIMERA). The peptide shown is that of p65 carrying the wt R41 and the mutants D41 and A41 along with some nearby residues. The NF-κB DNA binding site (10 bp) is shown in yellow and the site at equivalent position to that of the SNP rs28416813 is in red (12th bp). (Right) Equal amounts of plasmids encoding NF-κB p50 and either WT or mutant p65 were used as TFs to stimulate the IFN-λ3 promoter. The construct p1.4kbIFNL3 was used in the experiments with C or G alleles at the SNP rs28416813; the experiments were carried out on HEK293T cells grown in 96-well plates. The data is drawn from nine experiments representing three different plasmid preparations. The mean value from all nine experiments is shown, while the error bars depict SD. B) Overexpression of phosphomimic (constitutively active) versions of IRF3 and IRF7 and the mediator MAVS also reveals differences in transcriptional activity of p1.4kbIFNL3 carrying C or G alleles at rs28416813. The experiments were carried out on HEK293T cells grown in 96-well plates. The data is from three experiments performed with three different plasmid preparations. The mean value from the three experiments is shown, while the error bars depict SD. For both A and B, two tailed student t-test for independent means was used for calculating statistical significance; *p < 0.05, **p < 0.01; ***p < 0.001. All the Luciferase ratio values from each individual experiment are shown in Suppl. Table 1.