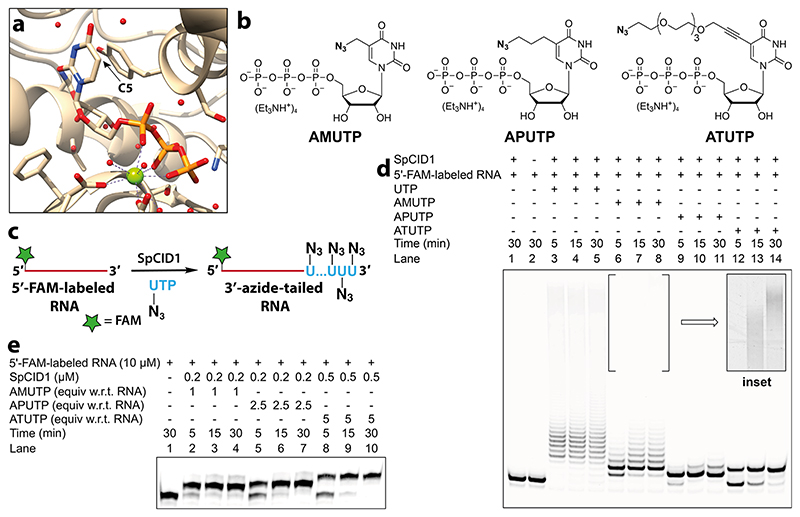
Figure 2. SpCID1, a TUTase enzyme, efficiently incorporates C5-azide-modified nucleotides at the 3’ end of an RNA oligonucleotide.
(a) Crystal structure of SpCID1 bound to UTP (PDB 4FH5) generated using UCSF Chimera software.46 C5 position of uridine triphosphate is shown using an arrow. (b) Structure of azide-modified UTP analogues used in terminal uridylation reaction. (c) Scheme showing 3’-terminal uridylation of model 5’-FAM-labeled RNA with modified UTP analogues. (d) Image of PAGE resolved reaction products of terminal uridylation reaction of 5’- FAM-labeled RNA (10 μM) in the presence of UTP/modified UTPs (500 μM) using SpCID1 (0.5 μM). Inset: Processive incorporation of AMUTP was evident upon adjusting the brightness and contrast of the gel image. See Figure S6 for the complete gel picture in these image settings. (e) Gel image showing optimized reaction conditions to effect single nucleotide incorporation.