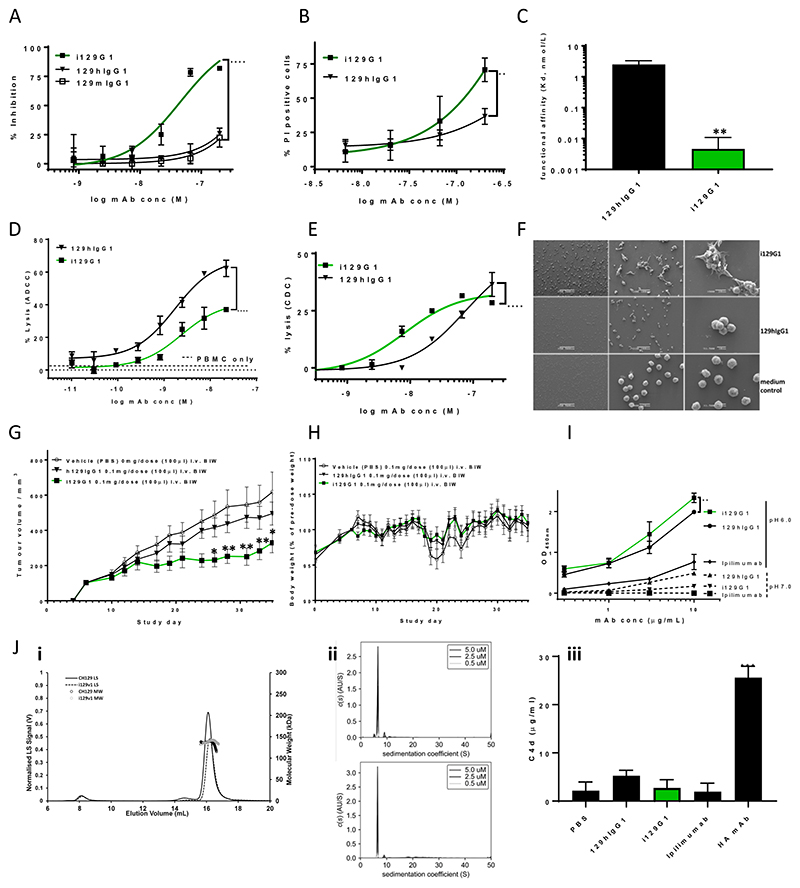
Figure 6. i129G1, derived from a non-cytotoxic mIgG1 mAb, exhibits significant direct cytotoxicity, enhanced avidity, pore forming ability as well as significant in vivo tumor control.
Significantly increased proliferation inhibition (A) and PI uptake (B) on COLO205 by i129G1 compared to 129hIgG1. Direct cell killing of low to moderate binding cancer cell lines was negligible (Supplementary Fig. 4). Significantly increased functional affinity (SPR) by i129G1 compared to 129hIgG1 (C). i129G1 maintains nanomolar ADCC activity on COLO205, but with significantly reduced overall lysis compared to h129hIgG1 (D). Nanomolar CDC activity by i129G1 on COLO205 is significantly increased compared to 129hIgG1 (E). Evidence of cellular detachment, aggregation and pore forming ability by i129G1 on COLO205 using SEM, white arrows point to irregular pores (F). Significant in vivo tumor control by i129G1 compared to vehicle control and compared to 129hIgG1 in a COLO205 xenograft model (Balb/c nude mice) (G). Individual tumor growth curves are shown in Supplementary Fig. 5. No significant effect on mean body weight during the course of the mouse study (H). Dose-dependent binding of rhFcRn by i129G1 and 129hIgG1 at pH6.0 (I). Significantly increased binding by i129G1 compared to 129hIgG1 at the top two concentrations. Negligible binding at pH7.0 by both constructs (I). Similar SEC-MALS profiles for i129G1 compared to 129hIgG1(Ji). A small increase in higher MW species is evident in i129G1 compared to 129hIgG1 via AUC (Jii). No significant increase in C4d generation upon incubation with human serum by i129G1, compared to 129hIgG1 (Jiii). Significance versus respective parental constructs was deduced from two-way ANOVA (direct cytotoxicity, effector functions, rhFcRn binding, in vivo tumor control) or one-way ANOVA (functional affinity and C4d detection), with Dunnett’s corrections for multiple comparisons.