Abstract
Inexpensive genotyping methods are essential to modern genomics. Here we present QUILT, which performs diploid genotype imputation using low-coverage whole genome sequence data. QUILT employs Gibbs sampling to partition reads into maternal and paternal sets, facilitating rapid haploid imputation using large reference panels. We show this partitioning to be accurate over many megabases, enabling highly accurate imputation close to theoretical limits and outperforming existing methods. Moreover, QUILT can impute accurately using diverse technologies, including using long reads from Oxford Nanopore Technologies, and a novel form of low-cost barcoded Illumina sequencing called haplotagging, with the latter showing improved accuracy at low coverages. Relative to DNA genotyping microarrays, QUILT offers improved accuracy at reduced cost, particularly for diverse populations that are traditionally underserved in modern genomic analyses, with accuracy nearly doubling at rare SNPs. Finally, QUILT can accurately impute (4-digit) HLA types, the first such method from low-coverage sequence data.
Introduction
Large genome-wide association studies (GWAS) are essential to modern human genomics. They pinpoint individual genes that contribute to specific phenotypes, allow for accurate measures of disease heritability, reveal genetic relationships between phenotypes, and allow for dissection of the contribution of tissues to specific phenotypes1. In addition, large GWASs are the basis to generate accurate polygenic risk scores2, which are essential to realizing the promise of precision medicine3.
Over the last decade, most GWASs have first genotyped half a million or more SNPs using a genotyping microarray, and then imputed additional untyped SNPs using haplotype reference panels4,5. The algorithms that perform the phasing and imputation are generally derived from the Li and Stephens model6, which models each sample individual as a mosaic of reference haplotypes. In such models, imputation accuracy increases with reference panel size, due to longer and hence more recent matches between sample and reference haplotypes, leading to increasingly large reference panels, for example the haplotype reference consortium (HRC)7. To handle these large panels, sophisticated phasing and imputation methods have been developed to work specifically from genotyping microarray input, resulting in fast run times and very high accuracy5,8–11.
Recently, low-coverage whole genome sequencing (lc-WGS) has emerged as an alternative to genotyping microarrays for obtaining genotyping information for imputation12–14. This approach has become increasingly attractive as both library costs15,16 and sequencing costs have decreased, and especially for non-model organisms, avoids expensive array design or low throughput costs17. Methods to impute from lc-WGS include some derived from approaches designed primarily for array based imputation18, with others designed specifically for lc-WGS17,19,20, or designed principally around non-human imputation21–23.
However, none of these methods are designed specifically for lc-WGS using large haplotype reference panels. Data from sequencing reads from lc-WGS have different properties than probes from genotyping microarrays, and warrant different statistical models for phasing and imputation. Firstly, probes from genotyping microarrays are short, being tens of base pairs long, and as such, information from different probes, covering different SNPs, is nearly always independent. By contrast, sequencing reads are typically 100-250 bases long, may be paired, and with long read sequencing, may be many thousands of bases long. As such, information at nearby SNPs may not be independent, by coming from the same read(s). This issue is particularly acute when reads are expected to span more SNPs on average, for example highly polymorphic regions like the major histocompatibility complex (MHC), in populations of species with high genetic diversity, and with long read sequencing technologies, such as Oxford Nanopore Technologies (ONT) and Pacific Biosciences. Secondly, information from genotyping microarrays is summed across paternal and maternal haplotypes, while sequencing reads come from either the paternal or maternal haplotype. If the maternal or paternal origin of each sequencing read can be determined, imputation becomes much simpler, because maternal and paternal reads can be separated and haploid imputation performed, which has linear or better computational complexity in reference panel size. This reduction of complexity can greatly speed up computation and ensures efficient processing of many thousands of samples in large studies.
Here we present QUILT, a method for rapid genotype imputation and phasing from lc-WGS using a large haplotype reference panel. QUILT uses efficient computational storage of reference haplotypes and an iterative two-step procedure with Gibbs sampling to efficiently impute samples from lc-WGS with linear computational complexity in the number of samples, SNPs and reference haplotypes. We evaluate QUILT’s performance versus genotyping microarrays on three exemplar data types, representing the diversity of sequencing approaches currently available: Illumina short read sequencing data; ONT, one example of long read sequencing data; and haplotagging data, a low-cost, scalable form of barcoded short-read (linked-read) sequencing, featuring partial sequencing of long (>100kb) haplotype segments. We evaluate QUILT’s ability to accurately partition reads and impute genotypes across these different data types using one individual sequenced on all three platforms (NA12878), and three larger additional datasets. We further demonstrate QUILT’s effectiveness in imputing across diverse populations from the 1000 Genomes Project, and QUILT’s ability to accurately impute human leukocyte antigen (HLA) Class I and Class II alleles. Finally, we investigate what these results mean for the relative merits of designing studies using lc-WGS, versus genotyping microarray technologies.
Results
Overview of model
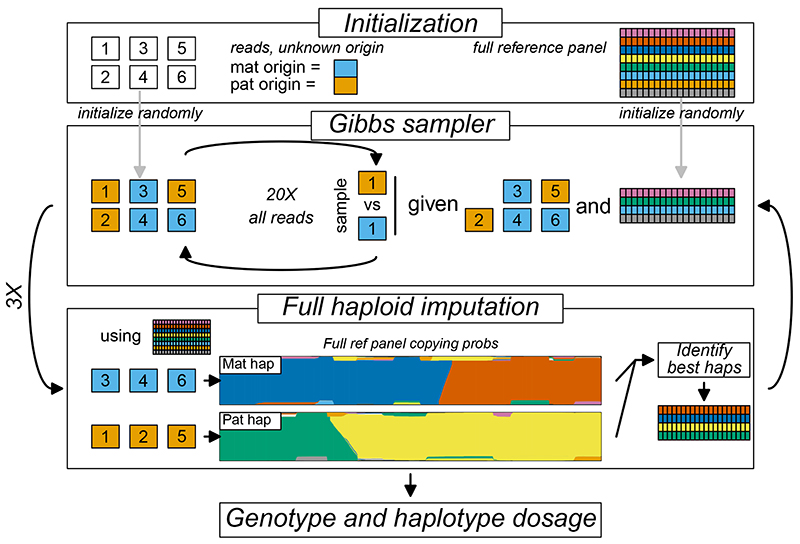
Our method, QUILT, models each sample haplotype to be imputed as a mosaic of a panel of reference haplotypes, using a Li and Stephens model6. Let K be the number of reference haplotypes. Because samples are diploid, a naïve implementation of this model has computational complexity that is proportional to K2, which is computationally prohibitive for large K. However, fundamentally, each sequencing read comes from either the maternal of paternal chromosome. Conceptually, if we could split them into two such sets, the imputation becomes computationally linear in K. Previously, in the STITCH model, we introduced a “pseudo-haploid” approach that had linear computational complexity17, by assigning approximating probabilities for each read to come from each background. Here, we improved this approach by implementing a Gibbs sampler through efficient re-use of stored forward-backward probabilities, allowing for re-sampling of all read labels into the two parental chromosomes, using a single forward-backward pass of the hidden Markov model (HMM). As Gibbs sampling is slow when using the full reference panel, we iterate between using the Gibbs sampler on a subset of the reference panel to update the read labels, and performing haploid imputation using the read labels and the full reference panel to update the subset of reference panel, and in the terminal iteration, perform the imputation (Figure 1). The Gibbs sampling is fast, by using a fraction of the panel, and accurate, as it uses the best matching reference haplotypes. Final imputation results are taken as an average across multiple Gibbs processes, and the entire algorithm is computationally linear in K. Further details, including phasing as well as computational efficiencies and speedups used in the model, are described in the Methods and Supplementary Note.
Figure 1. Schematic of QUILT model.
Model shown for one Gibbs sampling. Model is initialized for a vector of read labels, and a subset of reference haplotypes. The QUILT model then iteratively proceeds between Gibbs sampling, to obtain new read labels given the current subset of reference haplotypes, and full haploid imputation, to obtain new reference haplotype subsets using the current read labels. QUILT completes after a pre-specified number of iterations. Genotype dosage is taken as an average across Gibbs samplings, while phase is taken from an additional Gibbs sampling using read labels taken as average across previous samplings.
Imputation performance for NA12878
We obtained high coverage sequence for NA12878 using standard Illumina short read (“Illumina”)24, haplotagged Illumina short read (HT), and Oxford Nanopore Technologies long read sequence (ONT)25. We inferred accurate genotypes and haplotypes using high coverage trio data and phasing both using trio information and a haplotype reference panel (Methods), and use these as approximate truth haplotypes, from which we further generated probabilities that each sequencing read ultimately came from either the maternal or paternal haplotype (Methods). In what follows, we generally imputed 200 Mbp of the genome for comparisons, and for a reference panel, we used a subset of the haplotype reference consortium (HRC) dataset with NA12878 removed (therefore using 54,328 haplotypes)7 (Methods). In later sections we use different subsets of the HRC, after removing exact matches to test sequences. We chose parameter settings for QUILT that gave a reasonable trade-off between accuracy and speed across a wide range of coverages (Supplementary Figure 1).
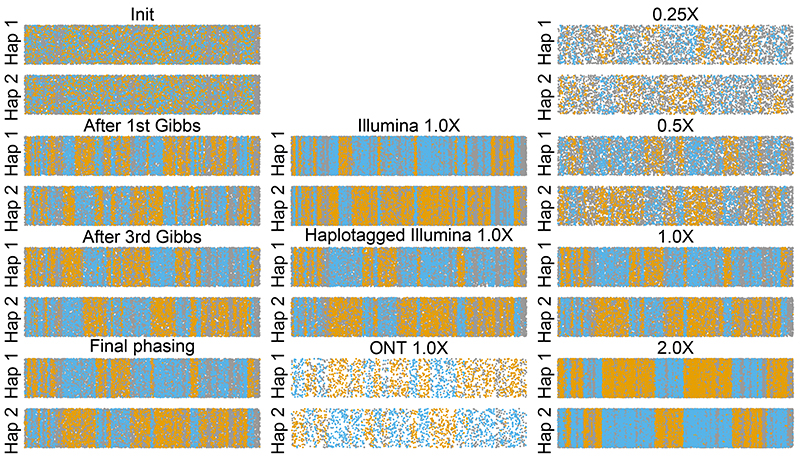
We assessed phasing accuracy both visually by comparing the continuity of inferred to truth read labels, and quantitatively using phase switch error rate. QUILT is able to achieve increasingly accurate read label phasing as the algorithm progresses, eventually achieving a read level phasing with approximately one switch error per megabase (Figure 2; errors are visualised as flips between runs of blue and orange, over a continuous 20 Mbp region of chromosome 20). When comparing across technology platforms, standard short-read Illumina data (unlinked) generated more switches than longer-read HT or ONT data (Figure 2). Indeed phase switch error rates were 0.09% for ONT, 0.08% for HT, and 0.13% for Illumina at 1.0X coverage (Supplementary Table 1). Phasing accuracy was robustly high across a wide spectrum of sequence coverage, increasing only to 0.11% (HT) at 0.25X coverage, though genotyping error rates increased as coverage decreased (5.26% of heterozygous sites at 0.25X, vs 2.12% at 1.0X, were erroneously imputed as homozygous).
Figure 2. Assessment of read label partitioning.
Per analysis, reads are grouped based on assignment to Hap1 or Hap2, with remaining y-axis variation being jitter. x-axis gives central location of read along 20 Mbp of chromosome 20. Reads are coloured blue and orange to reflect high posterior probability of coming from truth maternal or paternal chromosome, while grey indicates equally likely from either truth chromosome. Switches between runs of orange and blue denote probable switch errors. Columns denote effect of multiple iterations (left-most, for haplotagged 1.0X), different technologies (center, for 1.0X), and coverages (right-most, for haplotagged).
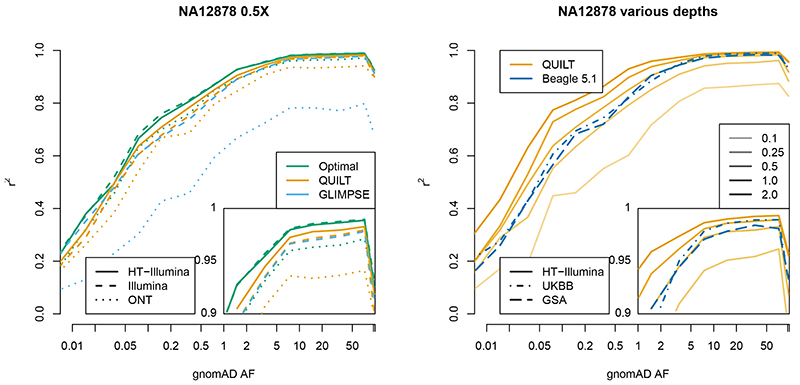
We next assessed genotype imputation accuracy across data types, methods, and coverage (Figure 3A, Supplementary Table 2). To explore the contribution of phasing errors to genotyping errors, we supplied our “truth” read label origins to the QUILT framework to generate what we call “Optimal” imputation results (these are optimal in the sense of possessing idealised phase information, but still vary with read coverage). We stratified results by allele frequencies from the separate genome aggregation database (gnomAD) 26. Throughout the results, we use “rare” to refer to SNPs with allele frequencies 0.1-0.2%, and “common” to refer to SNPs with frequencies 20-50%. Encouragingly, imputation results for QUILT at 0.5X are quite close to Optimal results and show a benefit for HT, particularly at rare SNPs (Optimal, HT, Illumina respectively; rare r2 = 0.76, 0.709, 0.678; common r2 = 0.988, 0.980, 0.975). This benefit for HT remained at lower coverages, for example for rare SNPs at 0.1X (HT, Illumina respectively; r2 = 0.460, 0.416). Comparisons with ONT are complicated slightly by the higher error rate of this platform, and at 0.5X ONT showed similar accuracy to Ilumina at rare SNPs (rare r2 = 0.669), and somewhat worse accuracy for common SNPs (r2 = 0.937), indicating a trade-off between better phasing and lower accuracy of individual reads.
Figure 3. Imputation accuracy of NA12878 sample.
r2 per-bin is aggregated over SNPs with a given gnomAD allele frequency for a given technology, coverage and method.
We compared QUILT results to GLIMPSE20 for each analysis (Figure 3A, Supplementary Table 2). As GLIMPSE uses genotype likelihoods from a VCF for input, there is no gain in using linked-read HT data under GLIMPSE versus standard Illumina sequencing. At 0.5X, results for QUILT and GLIMPSE were similar, with QUILT HT being the most accurate (QUILT HT, QUILT Illumina, GLIMPSE Illumina respectively; rare r2 = 0.709, 0.678, 0.672; common r2 = 0.980, 0.975, 0.974). For ONT, accuracy was considerably lower with GLIMPSE than QUILT (QUILT ONT, GLIMPSE ONT; rare r2 = 0.669, 0.428; common r2 = 0.937, 0.771). Run time comparisons between both QUILT and GLIMPSE for a variety of reference panel sizes for both low (NA12878, N=3) and moderate (1000G, N=93) sample sizes confirmed that QUILT has approximately linear computational complexity in reference panel size while GLIMPSE has constant computational complexity in reference panel size for moderate sample sizes (Supplementary Figure 2). Both methods had low and approximately linear relationships between memory usage and reference panel size. Both QUILT and GLIMPSE include parameter settings that trade off accuracy versus run time. Therefore we re-ran both methods, across a range of parameter values varying from their defaults, for NA12878 (Supplementary Figure 3, N=3, with three data types), as well as Illumina for 1000 Genomes data (Supplementary Figure 4, N = 93, a more realistic scenario involving more samples). The results indicate (when considering both speed and accuracy) that for lower-coverage data (0.1X) QUILT is favoured over GLIMPSE, for all three data types but especially HT and ONT data. For moderate coverages (0.5X) the methods are similar, except for ONT, and for high-coverage data (1.0, 2.0X) GLIMPSE is favoured over QUILT for the Illumina and HT data types, while QUILT is clearly favoured for ONT data.
We next compared QUILT results to those obtainable from genotyping microarrays. We approximated microarray input using high coverage WGS restricted to the relevant array SNP site list. We examined performance versus the commonly used Affymetrix UK Biobank (UKBB) and Illumina Global Screening Array (GSA) arrays, imputing using Beagle version 5.18 (Figure 3B, Supplementary Table 3). Both arrays yield similar accuracies at both rare SNPs (UKBB, GSA respectively; r2 = 0.694, 0.683) and common SNPs (r2 = 0.989, 0.984). For rare SNPs, all three sequencing platforms already exceeded this accuracy at 1.0X coverage (HT, Illumina, ONT respectively; r2 = 0.776, 0.754, 0.741). For the best-performing platform (HT), even at 0.5X coverage imputation was at least as accurate as arrays (r2 = 0.709 for rare SNPs, r2 = 0.988 for common SNPs), demonstrating the utility of QUILT and lc-WGS.
Imputation performance across diverse sequenced samples
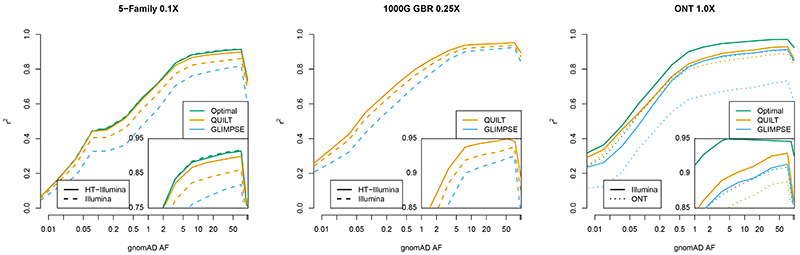
In three additional datasets, we confirmed the overall patterns we saw using NA12878 between data type and imputation performance. First, we further compared HT and Illumina using 7 offspring individuals from 5 different families of North American (N=3) and Ashkenazi Jewish (N=2) background (Figure 4A, see Supplementary Table 4 for all coverages). At 0.1X, we see the same ordering as with NA12878 for Optimal, QUILT HT, QUILT Illumina and GLIMPSE for both rare (r2 = 0.463, 0.450, 0.407, 0.328) and common (r2 = 0.911, 0.891, 0.850, 0.804) SNPs, although as coverage increased, the approaches became more similar. Second, we again compared HT and Illumina data using 59 samples of British background from 1000 Genomes (GBR, using 1000 Genomes24 notation) (Figure 4B, see Supplementary Table 5 for all coverages). At 0.25X, we see the same ordering for QUILT HT, QUILT Illumina and GLIMPSE for both rare (r2 = 0.629, 0.593, 0.519 respectively) and common (r2 = 0.947, 0.931, 0.917 respectively) SNPs. Finally, we further compared Illumina and ONT using 7 samples of diverse genetic backgrounds from Shafin et al.27 (Figure 4C, see Supplementary Table 6 for all coverages). At 1.0X, as for NA12878 imputation, we see that imputation from Illumina is more accurate than ONT, that QUILT is more accurate than GLIMPSE on ONT data, and similarly, within datatypes, we see the same relationship between Optimal, QUILT, and GLIMPSE, both for Illumina (rare r2 = 0.7, 0.64, 0.598; common r2 = 0.97, 0.925, 0.908 respectively) as well as for ONT (rare r2 = 0.638, 0.629, 0.439; common r2 = 0.905, 0.884, 0.718 respectively).
Figure 4. Imputation accuracy of 5-Family, GBR and ONT samples.
r2 per-bin is aggregate over all SNPs in that gnomAD allele frequency bin across all samples, for a given technology, coverage and method.
Imputation performance relative to genotyping arrays
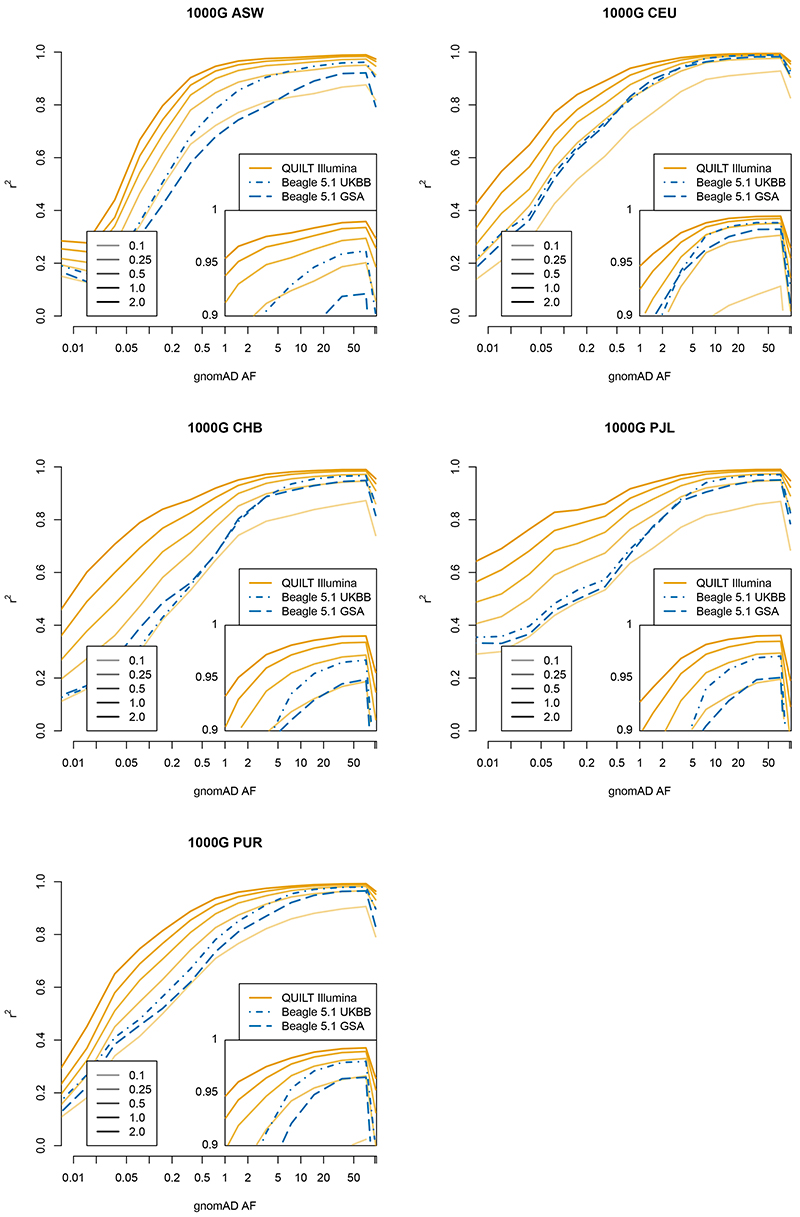
We next evaluated imputation performance versus genotyping microarrays (UKBB and GSA) across diverse samples using QUILT 1000G project samples for five groups (ASW, CEU, CHB, PJL and PUR) from distinct continental populations (Methods)24. As expected, imputation accuracy was highest for CEU (Northern and Western European) samples (Figure 5, Supplementary Table 7), who are most similar to the bulk of the HRC reference panel, where for rare SNPs, arrays were comparable to 0.25X sequencing data imputed using QUILT (r2 = 0.63-0.66). Interestingly for other groups, although absolute imputation quality declined, the relative performance of lc-WGS and QUILT increased compared to genotyping arrays. For example, for CHB (Han Chinese in Beijing) samples, lc-WGS and QUILT outperformed arrays for rare SNPs (QUILT, GSA, UKBB respectively; r2 = 0.581, 0.485, 0.43). For common SNPs imputation accuracy was generally high across platforms. Thus, the benefits of sequencing versus genotyping appear considerably greater for populations less similar to a given reference dataset. Higher coverage 1.0X and 2.0X lc-WGS data outperformed arrays at all frequencies, especially lower frequency variants, with QUILT nearly doubling the above array-based values (e.g. for rare SNPs, CHB, QUILT 1.0X r2 = 0.768, 2.0X r2 = 0.840).
Figure 5. Imputation accuracy of 1000 Genomes samples.
r2 per-bin is aggregate over all SNPs in that gnomAD allele frequency bin across all samples, for a given technology, coverage and method.
HLA imputation performance
Methods for accurate HLA imputation using genotypes28,29 or high-coverage sequence data30 have previously been developed. As part of QUILT, we developed a novel HLA imputation algorithm for lc-WGS, that uses reads inside an HLA locus for direct read mapping, and uses the remaining reads for imputation using a labelled reference panel (Online Methods). We used reference HLA data from both the HLA database IPD-IMGT/HLA31 and 1000 Genomes Project, and used withheld 1000 Genomes data for testing. We imputed HLA types for five populations (ASW, CEU, CHB, PJL and PUR) at each of 5 classical HLA loci (HLA-A, HLA-B, HLA-C, HLA-DQB1, HLA-DRB1). In addition, we assessed the accuracy of array-based HLA imputation using the same reference data and a method derived from the approach of SNP2HLA (Methods)28,32.
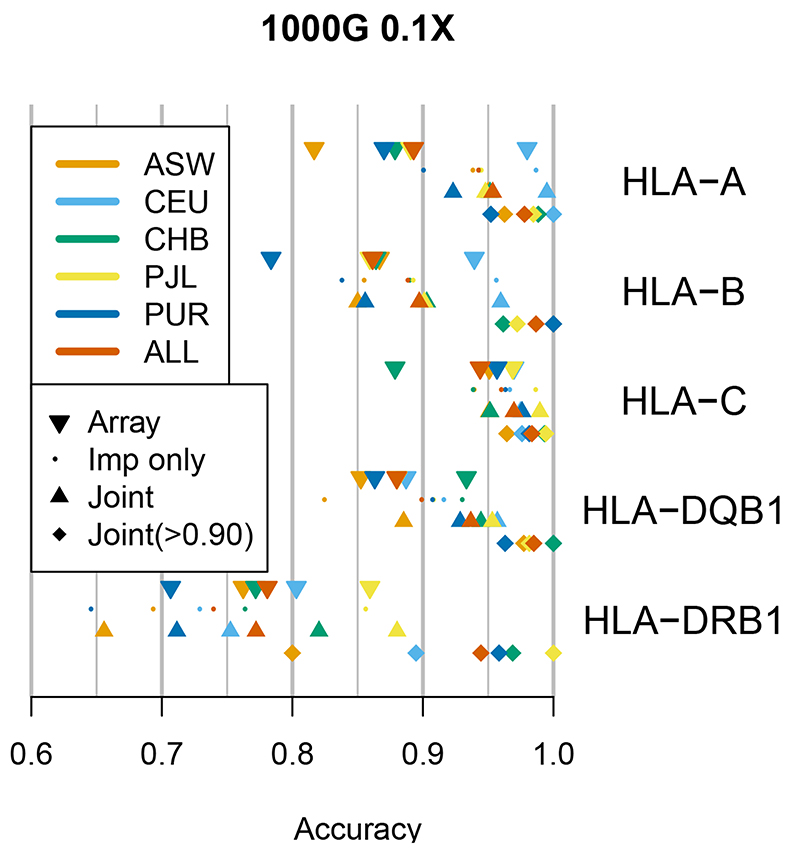
The results demonstrate that accurate HLA imputation from low-coverage sequence data is achievable across both Class I (HLA-A, HLA-B and HLA-C) and II (HLA-DQB1 and HLA-DRB1) loci, with the former showing generally higher accuracies (Figure 6, Supplementary Table 8). Using HLA-A as an example, the array based approach had an accuracy of 0.893 across all populations, while at 0.1X QUILT achieved 0.953 (0.995 CEU, 0.950 ASW (Americans of African Ancestry in SW USA)). Accuracy rose to 0.978 when only considering confidently called alleles. Accuracy decreased to 0.946 when the direct read information was not used, indicating that even at 0.1X direct read mapping can be useful. Results were consistent across coverages (Supplementary Figure 5), although direct read mapping provided a relatively larger boost in accuracy at higher coverage (9.3% at 2.0X) versus lower coverage (1.9% at 0.1X). Results for the other Class I loci of HLA-B and HLA-C, as well as HLA-DQB1 were similar, though HLA-B was less accurate across all samples, again mirroring known results29. Accuracies for HLA-DRB1 were lower, and this was the only locus where the array-based accuracy (0.781) slightly exceeded QUILT for 0.1X data (0.772). Nonetheless, accuracy at confidently called alleles remained high for QUILT at 0.1X, being 0.985 for HLA-DQB1, and at least 0.944 for every other locus. As expected, across all analyses, imputation accuracy was generally higher for more common HLA alleles, than rare alleles (Supplementary Table 8). Finally, we examined results for 7 specific HLA alleles chosen because they are either disease associated or associated with adverse drug reactions, similarly to previous work29. Except for some DRB alleles, accuracy at these medically important alleles was generally very high (Supplementary Table 9).
Figure 6. Imputation accuracy of HLA loci.
Accuracy is percent of correct unphased HLA alleles versus computationally inferred truth. Results are shown both per-population and in aggregate (ALL). Results are given both using only imputation (Imp only), as well as imputation plus direct read mapping (Joint, the default QUILT output). Results are further given at the subset of individuals with confidently inferred alleles (Joint(>0.90)). As reported elsewhere29, HLA Class I loci (HLA-A, HLA-B and HLA-C) are less diverse than Class II loci (HLA-DRB1 and HLA-DQB1) and thus yield more accurate imputation results.
Relative effective sample size and power
Finally, we performed a cost/benefit analysis of lc-WGS and QUILT-based imputation, versus widely used genotyping microarrays. We assessed the benefit in both a GWAS and a burden test setting, where for the latter the focus is on testing for an excess of rare coding variants in cases at a specific locus (Methods). We tested a scenario of using samples drawn from the CHB population, using estimates of imputation accuracy from the 1000 Genomes CHB analysis, and assumed a fixed cost of 30 GBP per array, and library costs from Meier et al.16. We varied both the cost of phenotyping a sample (i.e. non-genotyping costs), as well as the per-X sequencing costs, from approximate costs available today of 1000 USD / 30X genome, to potential lower future costs of 250 USD / 30X genome.
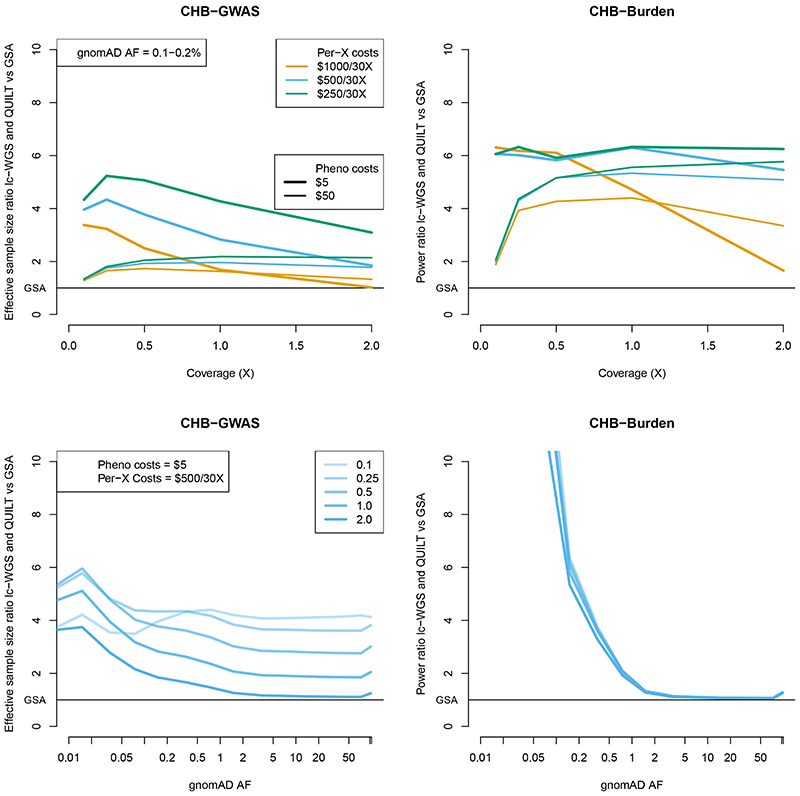
Results show that for both settings, sequencing yields nearly uniformly larger effective sample sizes or greater power than genotyping arrays (Figure 7, Supplementary Table 10, Supplementary Figure 6). For GWAS testing, lc-WGS and QUILT yield effective sample sizes 3.5 and 1.4 times larger than using genotyping microarrays for the inexpensive and expensive phenotyping cost, respectively, for identifying associations with rare SNPs of 0.1-0.2% frequency. For the burden test setting, power differences are even more pronounced. The relative gain in effective sample size or power increases as sequencing and phenotyping costs decrease. Results for fixed phenotyping and sequencing costs show the improvement of using lc-WGS and QUILT are especially pronounced at the lowest coverages. At current sequencing costs, the most cost-effective sequencing coverage is low, at less than 1X coverage, though imputation using higher coverage will be more useful in the future as sequencing costs decrease.
Figure 7. Relative increase in effective sample size and power using lc-WGS and QUILT.
Results are shown as a ratio of effective sample size for the GWAS setting, and a ratio of power for the burden test setting. Results use 1000 Genomes CHB imputation accuracy. Results for the top panel are given as a function of coverage, with variable phenotyping and per-X sequencing costs, for a fixed allele frequency (0.1-0.2%). Results for the bottom panel are given as a function of allele frequency, with varying coverage, assuming fixed phenotyping ($5 / sample) and per-X sequencing costs ($500 / 30X). All results assume a library preparation cost of 1.36 GBP /sample and an array cost of 30 GBP / sample.
Discussion
Inexpensive and accurate genotyping solutions are needed to perform the next generation of GWAS. In this work, we showed that QUILT can unlock the power of lc-WGS for this task, simultaneously improving accuracy and reducing costs for diverse data types and individuals from varied populations, compared to traditional genotyping arrays. Here we consider why we observed these results, the broad implications of the results observed here for GWAS and other studies, and finally natural extensions of this work.
To begin, it is helpful to consider imputation from lc-WGS, supposing we knew the true read label assignment, i.e. whether each sequencing read came from the maternal or paternal chromosome. Neglecting the impact of errors at rates <1% for either kind of data, standard arrays type on the order of a million SNPs across the genome, typically favouring common variants, and/or higher information content. For comparison, lc-WGS sequencing at 2.0X coverage - if we know the true read label assignment - yields about 1.0X coverage for each haplotype, or about 63% of the entire genome for each haplotype under a simple Poisson model of sequencing coverage. While information from some array SNPs is lost, information from many more non-array SNPs is gained, explaining why QUILT can be more accurate than arrays at higher coverage, particularly for populations other than those for which genotyping microarrays are principally designed for. The above relies on accurate read label assignments, and in practice, while QUILT does make some errors, assignment is highly accurate, explaining why QUILT results are often near “Optimal” values, where direct phase information is provided. This explains why short-read (Illumina-based) sequencing outperforms long-read data (exemplified by ONT), because the slightly improved read label assignment possible from long reads is outweighed by the much lower per-base error rate of the short-read data. The haplotagging approach, meanwhile, improves accuracy versus unlinked Illumina reads as it retains the same per-base error rate, but decreases phasing errors, particularly at low coverages, where there still remains more phasing uncertainty.
What do these results mean for GWAS today, and in the future? lc-WGS already provides similar accuracy to genotyping microarrays at only 0.1-0.25X coverage, and much higher accuracy at 1.0-2.0X. Combined with estimates of library and sequencing costs, in terms of power for a given expenditure, it is already preferable to use lc-WGS and QUILT as opposed to genotyping microarrays across a diverse range of scenarios. This is particularly true for discovering rare disease-associated variants, which recent work suggest harbour a disproportionate amount of heritability for human phenotypes33,34. Because QUILT, in contrast to other methods for lc-WGS, models reads directly, and can work with high per-base error rates, it is robust across sequencing technologies, ensuring accuracy will remain high regardless of future technological advances. Further, by modelling reads directly, QUILT should work well regardless of the diversity of the region of the genome, or in non-human species where heterozygosity levels are often higher, and can be an order of magnitude higher, than in most human populations16. The strong performance of QUILT for HLA imputation, across diverse sample backgrounds, offers a concrete confirmation of this prediction. QUILT has linear computational complexity in sample size and haplotype reference panel size, ensuring reasonable run times across a range of study sizes.
In terms of future development of the model, as datasets with millions of lc-WGS become available, they will provide extensive information to improve imputation, and we intend to address this in future work. For example, we might iterate imputation, followed by incorporation of imputed sequenced samples into the reference panel. This might offer particular improvements, for example for newly discovered rare variants not represented in the reference panel. The QUILT model could also be adapted to novel settings. As one example, non-invasive prenatal testing (NIPT) using low coverage sequence is rapidly becoming the clinical standard of care35, and can generate the genotypes, both fetal and maternal, needed for GWAS36. The model presented here could readily be adapted to this data type, to allow separate imputation of the three haplotypes present in NIPT: the maternal transmitted and untransmitted, and paternal transmitted sequences. This would enable recovery of the maternal, fetal and partial paternal genomes, allowing determination of e.g. carrier status for genetic risk variants, and polygenic risk scores.
In conclusion, lc-WGS imputation using QUILT is flexible and accurate across a range of coverages and datatypes. It is particularly beneficial for imputing rare SNPs, and for imputing genotypes for human populations poorly represented by existing large haplotype reference panels, and should help empower a new wave of large, ethnically diverse GWAS.
Online methods
QUILT
In the QUILT model, imputation is performed through an iterative two step process. First, sequencing read labels, reflecting maternal or paternal origin, are updated using Gibbs sampling based on a subset of the full reference panel. Second, based on the read labels, the sequencing reads are split into two sets and separately imputed using the full reference panel, and this is used both to determine the best matching haplotypes for the next round of the Gibbs sampler, and in the terminal iteration, provides the imputed genotypes to be output. Here we offer more high-level detail on how this works, with full details offered in the Supplementary Note. The full details includes an explanation of a generative model under which reads would be simulated, detailed mathematics for both the Gibbs sampler and full haploid imputation, as well as a description of the phasing procedure, and an explanation of other parameter choices. Note that throughout the description of the model, the term “read” will refer to potentially discontinuous sequencing information from a single DNA fragment, either a random variable or observed, but known to come from the same underlying molecule: i.e. two reads from a read pair will be called a read, since they are observations from the same molecule.
Let Gen be the genotype for some arbitrary SNP, and E[Gen|O] be the expected genotype given the observed sequencing data. Let λ be the parameters of the model, for example the recombination rate. We can use random sampling to estimate the diploid genotype dosage as
We use Gibbs samplings to generate draws from H ~ P(H | O, λ). Let the vector h be our realized Gibbs sampled value at any point during the sampler. We begin by initializing h at random, i.e. for read indexed by v, hv is drawn equally from {1,2} for all v (where arbitrarily 1 = maternal, 2 = paternal). Let o be the realized observations for random variable O (sequencing read bases and base qualities). We assume that the bases of a sequence read reflect the haplotype being copied from in the haplotype reference panel - specifically, who is being copied from at the central point of the read. Suppose we consider a Li and Stephens model and a hidden Markov model (HMM), where transition probabilities depend on the recombination rate in the usual way, and emission probabilities for a state (copied reference haplotype) at a site depend on the reference haplotype sequence and observed sequencing reads whose central location is that site (by the assumption above, the observations (reads) are independent between sites, ensuring this is a valid HMM). From this, we can calculate P(O=o | H = h, λ) using the forward backward algorithm. Now suppose we want to sample a new value at read label hv for read v conditional on all other read labels. Let Hv be this random variable at this point in the Gibbs sampler, and let H-v be a random variable representing the remaining read labels. We therefore need to calculate
for l in {1,2}, and sample hv using this probability (where we note P(O = o, H = h| λ) = P(O=o | H = h, λ) P(H = h| λ) is easy to switch between as P(H = h| λ), the probability of the read labels, is 1/2 to the power of the number of reads). Naively calculating the above requires a new forward backward pass of the HMM. We are able to avoid a new forward-backward pass through efficient re-use of the forward and backward variables, and in fact re-sample all read labels under the Gibbs sampler using a single forward-backward pass. Details of this, and further details including a block Gibbs sampler, are given in the Supplementary Note.
Now, while the above is computationally linear in K, the reference panel size, it can be slow for large K. Therefore, we run the above Gibbs sampler using a reduced set of haplotypes, default 400. Further, the above HMM is based on the assumption that each read has a central location, and genotypes in sequencing reads are based on the reference haplotype copied at that central location. This assumption is less accurate for long read sequencing, and moreover, is irrelevant once read labels are known, as once reads are known to come from the same haplotype, membership of a sequenced base in a read no longer matters. This in effect changes the nature of the observations at a given site from sequencing reads to sequenced bases and their base qualities, and allows us to work with haplotype genotype likelihoods. Therefore, given read labels, we split the reads into sets reflecting maternal and paternal origin, generate haplotype genotype likelihoods, and perform full haploid imputation using the entire reference panel. We then use posterior state probabilities to update the subset of the reference panel used by the Gibbs sampler, as well as optionally output the genotype dosages. Full details of this are given in the Supplementary Note.
Datasets
NA12878
Both the NA12878 sample here and the further samples from the 1000 Genomes centre came from the New York Genome Center (NYGC) resequencing effort. We generated haplotagged NA12878 Illumina short read data, and by either considering or ignoring BX tags, used this both for analyses involving haplotagging and those without. We used ~80X ONT data from Bowden et al.25, and converted it to GRCh38 by first downsampling it using samtools (version 1.0)37 to ~8X, converting to fastq using samtools, mapped using Minimap238 using --alt-drop 1.0 and used --alt with a list containing all non-autosomal, chrX, chrY or chrMT contigs, and sorted and indexed using samtools. Parental genomes were obtained from the Illumina Platinum Genomes project under ENA accession code PRJEB3381, and converted to GRCh38 per-chromosome for chromosomes 6, 20, 21, by first sorting reads by read name using samtools, then converting to fastq format using samtools, then read mapping using bwa mem (v0.7.15), then sorting, then results were combined using samtools cat, and finally re-sorted and indexed.
5-Family
We generated (9.4X to 16.4X coverage, mean 12.2X) haplotagged data both for imputation and for truth data for 7 offspring and 10 parents in 5 families from the Bloom Syndrome Repository (4 trios and one 2 parent plus 3 offspring family). Parental samples were similarly haplotagged (2.8-9.8X, mean 6.5X), though this information was not used for inference of “true” genotype or phasing. Samples were North American, and also include individuals of Ashkenazi Jewish origin. Written informed consent for all 5-Family individuals was obtained by the Institutional Review Board of Weill Cornell Medical College.
GBR
We generated low coverage (mean 0.30X, range 0.13X-0.47X) haplotagged data for the 91 GBR 10000 Genomes project samples, as described in the Haplotagging section of the Supplementary Note. We analysed the 59 samples with assessed depth greater than or equal to 0.25X. We used “truth” data for these samples from the 1000 Genomes project resequencing data from the New York Genome Centre24.
Shafin et al. Oxford Nanopore Technologies (ONT)
We downloaded high coverage ONT and 10X Illumina data from Shafin et al. for 7 samples27 HG01243, HG02080, HG03098, HG01109, HG02055, HG02723, HG03492 from Amazon’s pangenomics resource (https://s3-us-west-2.amazonaws.com/human-pangenomics/). We downloaded high coverage Illumina 1000 Genomes data for their parents from the NYGC. For the ONT data, we processed the first 20 million reads from the fastq file using Minimap2 (v2.1), in the same way as the NA12878 sample. For 10X Illumina data, we used the proc10xG toolkit (https://github.com/ucdavis-bioinformatics/proc10xG, commit 7afbfcf), which successively runs process_10xReads.py, bwa mem, samConcat2Tag.py and samtools view -b to generate a mapped bam file.
1000 Genomes data
We used high coverage 1000 Genomes project resequencing data from the New York Genome Centre24. We chose one population (ASW, CEU, CHB, PJL, PUR) from each of the 5 continental superpopulations within 1000G (AFR, EUR, EAS, SAS, AMR). To minimize computation time for full imputation we chose every 5th member of this subset to impute, resulting in N=11 ASW, N=23 CEU, N=19 CHB, N=20 PJL, N=20 PUR samples. Due to a lack of consistent parent or offspring high coverage genome availability, we did not phase the data. For imputation of HLA loci we used all members of each population, and we generated and applied a modified reference panel, as described in the HLA imputation methods section.
Mapping and downsampling
All mapping was done against the human reference genome GRCh38, and when performed using bwa mem used (at least) options -Y -K 100000000. Average depth was calculated using samtools depth -a -q 10 -Q 10 on chromosome 20 from 1 to 10 Mbp for each BAM. Reads were grouped into molecules, either using paired read information, or using the BX tag from haplotagging. Downsampling was performed per-molecule (i.e. all reads from both read pairs and given linkage information from the BX tag were taken as being from the same molecule) using a Bernoulli probability determined by the desired coverage. For the 5-Family samples, we used results from an earlier version of the chemistry for haplotagging, as a later re-run achieved less than 2X coverage for each sample. We then considered the proportions of molecules that had no BX tag, had 2 or fewer reads in a BX tag, or had 3 of more reads in a BX tag. When downsampling using data from the old chemistry, we also downsampled to ensure these proportions matched results from the new chemistry. Subsampled BAMs were written either using the original read names (QNAME field of the BAM) or using new reads names that incorporated the BX molecule information.
Reference panel
We used a controlled access version of the HRC7 with Genbank accession number EGAD00001002729. We used liftOver to convert the reference panel from the GRCh37 to the GRCh38 reference genome using GATK Picard LiftoverVCF (v2.22.2)39. From the original GRCh37 HRC reference panel (39,131,578 autosomal variants), we removed 6,803 variants due to failure to map between the GRCh37 and GRCh38 reference genomes and a further 9,010 variants due to mismatching chromosome between the two reference genomes. The resulting autosomal GRCh38 HRC reference panel contain 39,115,765 variants and 27,165 samples with 54,330 haplotypes.
For imputation, we created three versions of the reference panel, based on three different subsets of the full panel. We first made a subset where we removed only NA12878, and this was used for the analyses involving NA12878 and the 5-Family individuals. Secondly, we made a version where we removed NA12878, the parents on the ONT samples from Shafin et al. used in this study, and also removed those samples from the 5 populations of 1000 Genomes we used for testing imputation (ASW, CEU, CHB, PJL, PUR), and used this on the ONT and 1000 Genomes analyses. Thirdly, we made a version where we removed NA12878 and the entire GBR population, for the analyses involving the GBR dataset.
Imputation
We used QUILT v0.1.340, GLIMPSE20 version 1.0.0 (github commit 00a0c55a2c23c8f47c832647a5a5e6806bc69802), and Beagle 5.18 (beagle.18May20.d20.jar), using default parameters across all runs. All imputation was done in 2 Mbp chunks, with 500kbp flanking buffers for all methods, on three regions: chr6:1-172000000bp, chr21:1-26000000bp, and chr21:14000001-46000000bp. QUILT utilises a pre-constructed compressed internal data format derived from the relevant .hap and .legend format files. For QUILT and GLIMPSE, we used a CEU based recombination map (CEU_omni_recombination_20130507.tar), and liftOver to generate a GRCh38 recombination rate map, across all runs. For Beagle, we used a plink format recombination rate map available from the Beagle web site (plink.<chr>.fixed.GRCh38.map). For GLIMPSE, we used GATK 3.8-1-0-gf15c1c3ef UnifiedGenotyper --genotyping_mode GENOTYPE_GIVEN_ALLELES --output_mode EMIT_ALL_SITES, to generate genotype likelihoods at sites to impute, which was done using GLIMPSE_phase. For both QUILT and GATK as input for GLIMPSE, we used a minimum base quality and mapping quality of 10 (using -minBQ in GATK). For input to Beagle representing arrays, we intersected the high coverage truth genotypes called using GATK UnifiedGenotyper with sites present on the array, to generate an input VCF. To determine sites, for the UK Biobank Affymetrix array, we used an annotation file (Axiom_tx_v1.na35.annot.csv), removing a small number of array sites that did not map to the autosomes or sex chromosomes, were not unique, or were multi-allelic, and used liftOver to get results in GRCh38, yielding 761,888 SNPs. For the Illumina Global Screening Array (GSA), we used strand files from https://www.well.ox.ac.uk/~wrayner/strand/, specifically GSA-24v3-0_A2-b38.strand.
Assessing imputation and phasing accuracy
We called “truth” genotypes using high coverage Illumina short read bams at HRC bi-allelic SNPs using the GATK UnifiedGenotyper software and option “—alleles” at the bi-allelic SNPs, setting genotyping_mode GENOTYPE_GIVEN_ALLELES and output_mode EMIT_ALL_SITES39. We used truth genotypes from high coverage sequencing only at sites where depth was at least 6. Phasing of high coverage trios was done first assuming Mendelian inheritance, excluding triple-heterozygous sites, using bespoke R code, then the excluded sites phased using this scaffold using shapeit411 (version 4.0 conda installation) using the HRC reference panel with only NA12878 removed.
To assess genotype imputation accuracy, we used imputed (test) dosages and high coverage (truth) genotypes. We assessed results either per-sample or across samples for SNPs in a given frequency range by constructing vectors of test dosages and truth genotypes and taking their squared Pearson correlation (in R using “cor”) at pairwise complete sites (i.e. ignoring truth sites with depth less than 6). SNP frequencies were taken from the gnomAD26 version 3.0 release, while for the very small number of SNPs in HRC not in gnomAD we used their HRC allele frequencies. We downloaded gnomAD data from https://gnomad.broadinstitute.org/downloads using links exemplified like the following for chromosome 20: https://storage.googleapis.com/gnomad-public/release/3.0/vcf/genomes/gnomad.genomes.r3.0.sites.chr20.vcf.bgz
To assess phasing accuracy we used switch error rates as follows. First consider we have both “truth” integer haplotype data and test imputed haplotype dosages (i.e. two real numbers within the range of 0 to 1), at sites where the truth genotypes are heterozygous. Define as discordant any test sites that are also not heterogyzous (sum of haplotype dosages does not round to 1). On the remaining sites, define a phase switch error as when either the truth haplotypes record a change in which haplotype carries the alternate allele between adjacent heterozygous sites when the test haplotypes do not, or vice versa. We removed from consideration sites that were flipped, i.e. yielding consecutive phase switch errors. The phase switch error rate is the number of phase switch errors divided by the total number of pairs of consecutive heterozygous sites examined, and can be combined across discrete imputed windows.
HLA imputation
We used HLA reference data built as follows. In brief, we downloaded full-length HLA alignments for annotated HLA genes and pseudogenes from the HLA database IPD-IMGT/HLA31 (version 3.39). This provides a set of (aligned) sequenced alleles for each region, to which reads can be mapped. Separately, for each HLA region, we subsetted reference haplotypes from the HRC to samples from the 1000 Genomes Project24. We then obtained unphased HLA types for those samples (version 20181129), previously inferred using high-coverage exome sequence data and using the PolyPheMe software41, and previously shown to have high accuracy. We phased HLA types onto haplotypes using a bespoke approach, and then excluded all members of the test populations, and those individuals with unphased alleles, from the labelled reference panel. For full details, see the Supplementary Note.
For HLA imputation, we used QUILT-HLA v0.1.6 for imputation from lc-WGS. For array based imputation, we were unable to run either SNP2HLA or the SNP2HLA module of HLA-TAPAS without error, so we used a custom HLA imputation approach implementing the algorithm used by SNP2HLA28,32. In brief, we first generated genotypes simulating an array, using high coverage WGS data at array sites, then used the HRC to impute additional sites from those genotypes using Beagle 4.18, and then finally used the labelled haplotype reference panel and the imputed genotypes to impute HLA types using Beagle 4.1. For details, see the Supplementary Note.
Cost effectiveness
We assessed the relative cost benefits of lc-WGS and genotyping microarrays using imputed results for rare SNPs (0.1-0.2%) and common SNPs (20-50%) for the CHB 1000 Genomes imputation results. We assumed a fixed array cost of 30 GBP per array, library cost of 1.36 GBP16, and per-X sequencing costs from 1000, 500 and 250 USD / sample, i.e. per-X costs of 1000/30, 500/30 and 250/30, converting into GBP using an exchange rate of 0.79375. For the GWAS type analysis, for a fixed budget, we calculate the number of samples available for imputation as budget / (pheno_cost + librar_cost + per_x_cost x coverage), while for genotyping microarrays, the number of samples is budget / (pheno_cost + cost_array). We then take the effective sample size as the sample size multiplied by the imputation r2, and take the ratio of these to be the relative increase in effective sample size from using lc-WGS.
For the burden style analysis, we assume a gene with 10 causal SNPs each possessing a frequency of 0.1% in cases and 0.01% in controls, and use the same imputation r2, i.e. the imputation r2 for SNPS at frequency 0.01% in the population. For simplicity, we may approximate this as 1 causal SNP with a frequency of f 1=1% in cases and f 2=0.1% in controls. We then suppose error is introduced governed by some parameter m, such that in cases, where X is the true genotype, and Y is the observed genotype (with imputation error), that P(X = 0, Y = 0) = 1 - f1 - m f1, P(X = 0, Y = 1) = m f1, P(X = 1, Y = 0) = m f1, P(X = 1, Y = 1) = f1 (1 - m), and for controls, that P(X = 0, Y = 0) = 1 - f2 - m f1, P(X = 0, Y = 1) = m f1, P(X = 1, Y = 0) = m f2, P(X = 1, Y = 1) = f2 (1 - m). We can then calculate m given r2. We then estimated power using 10,000 simulations using Fisher’s Exact test, given an alpha of 0.05 / 20000 (approximately Bonferroni-correcting for the number of genes in the genome), and given equal case and control numbers governed by the same N as for the GWAS analysis, where the budget was 10000 x (30 + pheno_cost), i.e. the default budget was the array cost plus phenotyping cost.
Supplementary Material
Acknowledgements
We thank Christa Lanz, Rebecca Schwab, Oliver Weichenrieder and Ilja Bezrukov at the MPI Developmental Biology for assistance with high-throughput sequencing and associated data processing; Andre Noll and the MPI Tübingen IT team for computational support. We used high coverage resequencing of 1000 Genomes project data performed by the New York Genome Center. These data were generated at the New York Genome Center with funds provided by NHGRI Grant 3UM1HG008901-03S1. The research was supported by the Wellcome Trust Core Award Grant Number 203141/Z/16/Z with additional support from the NIHR Oxford BRC, and by Wellcome Trust Grants 200186/Z/15/Z and 212284/Z/18/Z (to S.M.). The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health. We would also like to acknowledge the contribution and support from affected persons and their families who contributed to the Bloom Syndrome Repository. We thank the New York Community Trust and Weill Cornell Medicine’s Clinical and Translational Science Center for providing funding. M.K., D.S. and Y.F.C. are supported by the Max Planck Society and a European Research Council Starting Grant (No. 639096 “HybridMiX”).
Footnotes
Author contributions
R.W.D. developed and implemented QUILT. M.K, D.S. and Y.F.C. developed haplotagging. R.W.D, M.K and S.S. performed analyses. M.F. and C.M.C. developed the 5-Family dataset. S.M. developed and implemented the QUILT HLA typer. R.W.D, Y.F.C. and S.M. wrote the paper. All authors reviewed and approved the final manuscript.
Competing interests
M.K. and Y.F.C. declare competing financial interests in the form of patent and employment by the Max Planck Society. The remaining authors declare no competing interests.
Data availability
HRC haplotypes are available under accession EGAD00001002729 described at https://www.ebi.ac.uk/ega/datasets/EGAD00001002729 and are available through the Sanger Institute under controlled access.
High coverage whole genome sequence from the 1000 Genomes New York Genome Center collection was available through the links provided on this website https://www.internationalgenome.org/data-portal/data-collection/30x-grch38
Specifically we used the links from this file http://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data_collections/1000G_2504_high_coverage/1000G_2504_high_coverage.sequence.index
High coverage ONT from Bowden et al. is available through the ENA under accession PRJEB30620 from https://www.ebi.ac.uk/ena/data/view/PRJEB30620
High coverage ONT and Illumina (10X) samples from Shafin et al. are available through https://s3-us-west-2.amazonaws.com/human-pangenomics/index.html?prefix=
gnomAD SNP frequencies from the version 3.0 release were downloaded as detailed on https://gnomad.broadinstitute.org/downloads from URLs such as https://storage.googleapis.com/gnomad-public/release/3.0/vcf/genomes/gnomad.genomes.r3.0.sites.chr1.vcf.bgz
IPD-IMGT/HLA data was downloaded through their github database (https://github.com/ANHIG/IMGTHLA), specifically version 3.39 through URL https://github.com/ANHIG/IMGTHLA/blob/032815608e6312b595b4aaf9904d5b4c189dd6dc/Alignments_Rel_3390.zip?raw=true
Previously inferred HLA types for 1000 Genomes Project participants (version 20181129) were downloaded from the 1000 Genomes FTP http://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data_collections/HLA_types/20181129_HLA_types_full_1000_Genomes_Project_panel.txt
Recombination rates for CEU 1000 Genomes Project samples were downloaded from ftp-trace.ncbi.nih.gov/1000genomes/ftp/technical/working/20130507_omni_recombination_rates/CEU_omni_recombination_20130507.tar
All novel high and low coverage sequencing done for this study are available at NCBI’s short-read archive under BioProject accession PRJNA669554.
Code availability
QUILT is available from https://github.com/rwdavies/QUILT under a GPL license. The specific versions of QUILT used in this manuscript are available from Figshare40.
References
- 1.Pasaniuc B, Price AL. Dissecting the genetics of complex traits using summary association statistics. Nature Reviews Genetics. 2017;18:117–127. doi: 10.1038/nrg.2016.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dudbridge F. Power and Predictive Accuracy of Polygenic Risk Scores. PLOS Genetics. 2013;9:e1003348. doi: 10.1371/journal.pgen.1003348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Torkamani A, Wineinger NE, Topol EJ. The personal and clinical utility of polygenic risk scores. Nature Reviews Genetics. 2018;1 doi: 10.1038/s41576-018-0018-x. [DOI] [PubMed] [Google Scholar]
- 4.Burton PR, et al. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bycroft C, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li N, Stephens M. Modeling Linkage Disequilibrium and Identifying Recombination Hotspots Using Single-Nucleotide Polymorphism Data. Genetics. 2003;165:2213–2233. doi: 10.1093/genetics/165.4.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Consortium, the H. R et al. A reference panel of 64,976 haplotypes for genotype imputation. Nature Genetics. 2016;48:1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Browning BL, Zhou Y, Browning SR. A One-Penny Imputed Genome from Next-Generation Reference Panels. The American Journal of Human Genetics. 2018;103:338–348. doi: 10.1016/j.ajhg.2018.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Connell J, et al. Haplotype estimation for biobank-scale data sets. Nature Genetics. 2016;48 doi: 10.1038/ng.3583. ng.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loh P-R, Palamara PF, Price AL. Fast and accurate long-range phasing in a UK Biobank cohort. Nature Genetics. 2016;48 doi: 10.1038/ng.3571. ng.3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delaneau O, Zagury J-F, Robinson MR, Marchini JL, Dermitzakis ET. Accurate, scalable and integrative haplotype estimation. Nature Communications. 2019;10:5436. doi: 10.1038/s41467-019-13225-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pasaniuc B, et al. Extremely low-coverage sequencing and imputation increases power for genome-wide association studies. Nature Genetics. 2012;44:631–635. doi: 10.1038/ng.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Converge Consortium. Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature. 2015 doi: 10.1038/nature14659. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nicod J, et al. Genome-wide association of multiple complex traits in outbred mice by ultra-low-coverage sequencing. Nat Genet. 2016 doi: 10.1038/ng.3595. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rohland N, Reich D. Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Res. 2012;22:939–946. doi: 10.1101/gr.128124.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meier JI, et al. Haplotype tagging reveals parallel formation of hybrid races in two butterfly species. bioRxiv. 2020:2020.05.25.113688. doi: 10.1101/2020.05.25.113688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davies RW, Flint J, Myers S, Mott R. Rapid genotype imputation from sequence without reference panels. Nature Genetics. 2016;48:965–969. doi: 10.1038/ng.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Browning BL, Browning SR. Genotype Imputation with Millions of Reference Samples. Am J Hum Genet. 2016;98:116–126. doi: 10.1016/j.ajhg.2015.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spiliopoulou A, Colombo M, Orchard P, Agakov F, McKeigue P. GeneImp: Fast Imputation to Large Reference Panels Using Genotype Likelihoods from Ultra-Low Coverage Sequencing. Genetics. 2017 doi: 10.1534/genetics.117.200063. genetics.117.200063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rubinacci S, Ribeiro DM, Hofmeister RJ, Delaneau O. Efficient phasing and imputation of low-coverage sequencing data using large reference panels. Nature Genetics. 2021;53:120–126. doi: 10.1038/s41588-020-00756-0. [DOI] [PubMed] [Google Scholar]
- 21.VanRaden PM, Sun C, O’Connell JR. Fast imputation using medium or low-coverage sequence data. BMC Genetics. 2015;16:82. doi: 10.1186/s12863-015-0243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ros-Freixedes R, et al. Accuracy of whole-genome sequence imputation using hybrid peeling in large pedigreed livestock populations. Genetics Selection Evolution. 2020;52:17. doi: 10.1186/s12711-020-00536-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng C, Boer MP, van Eeuwijk FA. Accurate Genotype Imputation in Multiparental Populations from Low-Coverage Sequence. Genetics. 2018 doi: 10.1534/genetics.118.300885. genetics.300885.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.‘The 1000 Genomes Project Consortium’. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bowden R, et al. Sequencing of human genomes with nanopore technology. Nat Commun. 2019;10:1–9. doi: 10.1038/s41467-019-09637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karczewski KJ, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shafin K, et al. Nanopore sequencing and the Shasta toolkit enable efficient de novo assembly of eleven human genomes. Nature Biotechnology. 2020:1–10. doi: 10.1038/s41587-020-0503-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jia X, et al. Imputing Amino Acid Polymorphisms in Human Leukocyte Antigens. PLOS ONE. 2013;8:e64683. doi: 10.1371/journal.pone.0064683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karnes JH, et al. Comparison of HLA allelic imputation programs. PLoS One. 2017;12 doi: 10.1371/journal.pone.0172444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nature Biotechnology. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson J, et al. IPD-IMGT/HLA Database. Nucleic Acids Res. 2020;48:D948–D955. doi: 10.1093/nar/gkz950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo Y, et al. A high-resolution HLA reference panel capturing global population diversity enables multi-ethnic fine-mapping in HIV host response. medRxiv. 2020:2020.07.16.20155606. doi: 10.1101/2020.07.16.20155606. [DOI] [Google Scholar]
- 33.Durvasula A, Lohmueller KE. Negative selection on complex traits limits phenotype prediction accuracy between populations. The American Journal of Human Genetics. 2021;108:620–631. doi: 10.1016/j.ajhg.2021.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wainschtein P, et al. Recovery of trait heritability from whole genome sequence data. bioRxiv. 2019:588020. doi: 10.1101/588020. [DOI] [Google Scholar]
- 35.Snyder MW, et al. Copy-Number Variation and False Positive Prenatal Aneuploidy Screening Results. New England Journal of Medicine. 2015;372:1639–1645. doi: 10.1056/NEJMoa1408408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu S, et al. Genomic Analyses from Non-invasive Prenatal Testing Reveal Genetic Associations, Patterns of Viral Infections, and Chinese Population History. Cell. 2018;175:347–359.:e14. doi: 10.1016/j.cell.2018.08.016. [DOI] [PubMed] [Google Scholar]
- 37.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Genetics. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.QUILT source code from manuscript. figshare. 2021 doi: 10.6084/m9.figshare.14401904.v1. /articles/code/QUILT_source_code_from_manuscript/14401904/1. [DOI] [Google Scholar]
- 41.Abi-Rached L, et al. Immune diversity sheds light on missing variation in worldwide genetic diversity panels. PLOS ONE. 2018;13:e0206512. doi: 10.1371/journal.pone.0206512. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
HRC haplotypes are available under accession EGAD00001002729 described at https://www.ebi.ac.uk/ega/datasets/EGAD00001002729 and are available through the Sanger Institute under controlled access.
High coverage whole genome sequence from the 1000 Genomes New York Genome Center collection was available through the links provided on this website https://www.internationalgenome.org/data-portal/data-collection/30x-grch38
Specifically we used the links from this file http://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data_collections/1000G_2504_high_coverage/1000G_2504_high_coverage.sequence.index
High coverage ONT from Bowden et al. is available through the ENA under accession PRJEB30620 from https://www.ebi.ac.uk/ena/data/view/PRJEB30620
High coverage ONT and Illumina (10X) samples from Shafin et al. are available through https://s3-us-west-2.amazonaws.com/human-pangenomics/index.html?prefix=
gnomAD SNP frequencies from the version 3.0 release were downloaded as detailed on https://gnomad.broadinstitute.org/downloads from URLs such as https://storage.googleapis.com/gnomad-public/release/3.0/vcf/genomes/gnomad.genomes.r3.0.sites.chr1.vcf.bgz
IPD-IMGT/HLA data was downloaded through their github database (https://github.com/ANHIG/IMGTHLA), specifically version 3.39 through URL https://github.com/ANHIG/IMGTHLA/blob/032815608e6312b595b4aaf9904d5b4c189dd6dc/Alignments_Rel_3390.zip?raw=true
Previously inferred HLA types for 1000 Genomes Project participants (version 20181129) were downloaded from the 1000 Genomes FTP http://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data_collections/HLA_types/20181129_HLA_types_full_1000_Genomes_Project_panel.txt
Recombination rates for CEU 1000 Genomes Project samples were downloaded from ftp-trace.ncbi.nih.gov/1000genomes/ftp/technical/working/20130507_omni_recombination_rates/CEU_omni_recombination_20130507.tar
All novel high and low coverage sequencing done for this study are available at NCBI’s short-read archive under BioProject accession PRJNA669554.
QUILT is available from https://github.com/rwdavies/QUILT under a GPL license. The specific versions of QUILT used in this manuscript are available from Figshare40.