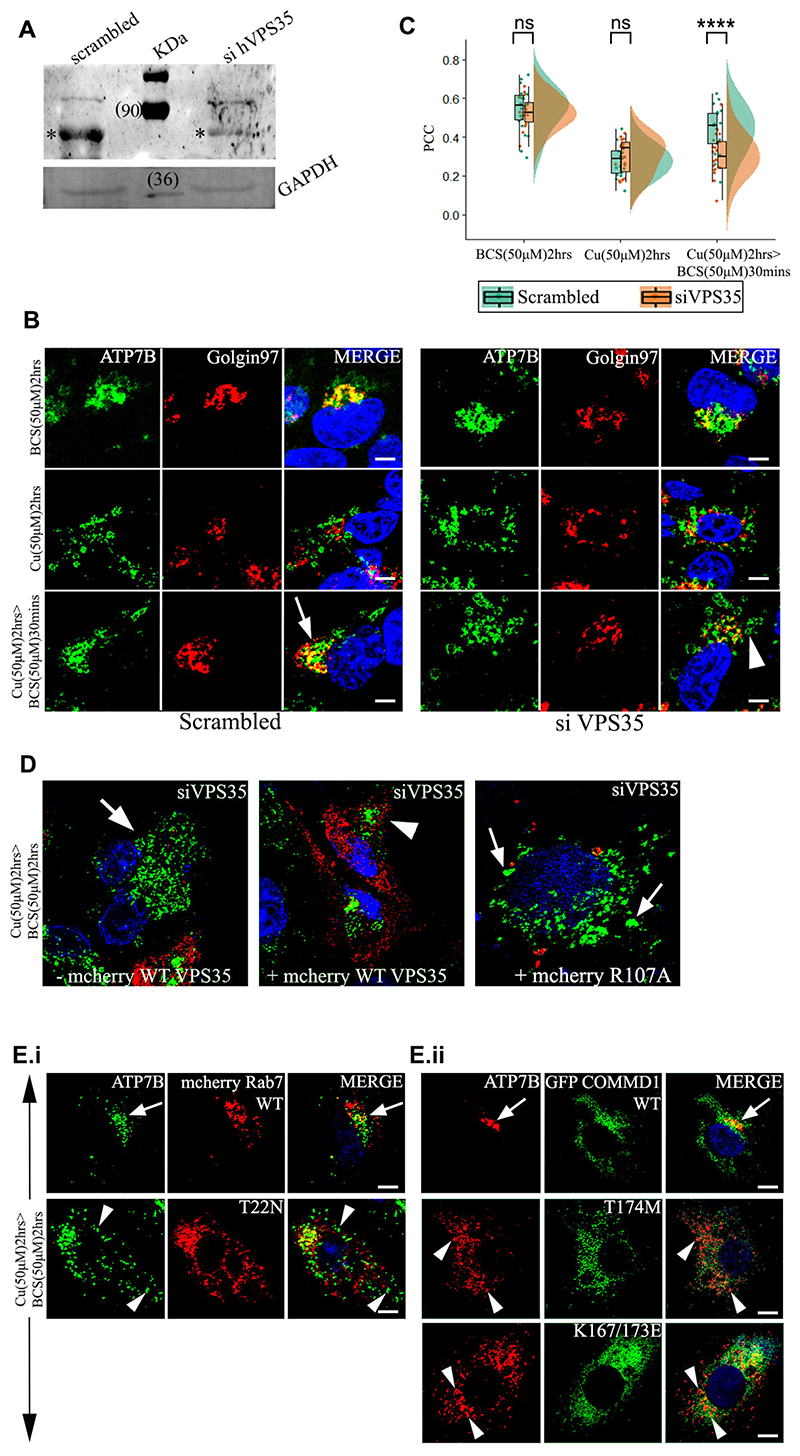
Fig. 4. VPS35 and its associated proteins regulates retrieval of ATP7B from lysosomes to TGN.
(A) siRNA-mediated knockdown of Vps35 in HepG2 cells shows its downregulation. (*) denotes the VPS35 protein. GAPDH is used as a loading control. (B) Colocalization of ATP7B (green) with the TGN marker Golgin97 (red) in BCS (top row) and 50 μM copper (second row) and copper depletion post copper treatment (third row). Arrow denotes TGN colocalization of ATP7B; arrowhead denotes vesicularized ATP7B. Scale bars: 5 μm. (C) Pearson’s correlation coefficient of colocalization between ATP7B and TGN at different copper conditions comparing VPS35 siRNA treated versus control demonstrated by a box plot. The box represents the 25–75th percentiles, and the median is indicated. The whiskers show the data points within the range of 1.5× interquartile range (IQR) from the 1st and 3rd quartile. ****P<0.0001; ns, not significant (non-parametric Mann–Whitney U test). (D) Localization of ATP7B (green) in VPS35 siRNA-treated cells and subsequently transfected with mCherry–wtVPS35 (red). The left image represents cells that are not expressing mCherry–wtVPS35 as compared to cells expressing mCherry–wtVPS35 (center image) or the mutant, mCherry-R107A-VPS35 (right) Cells belong to the same culture dish for both the left and center images. Arrow represents vesicularized ATP7B and arrowhead perinuclear ATP7B. (E.i.) Localization of ATP7B (green) in cells overexpressing mCherry–wtRab7 (red) (top panel) or dominant negative mutant mCherry-T22N-Rab7 (bottom panel) in copper to BCS condition to triggers retrograde trafficking of ATP7B. (E.ii.) At the same copper conditions as E.i., localization of ATP7B (red) in cells overexpressing GFP–wtCOMMD1 (top panel) or the mutants GFP–T174M-COMMD1 (middle panel) and GFP–K167/173L-COMMD1 (bottom panel). Scale bars: 5 μm. Blue signal represents DAPI staining for nucleus. Arrows represents perinuclear ATP7B and arrowheads vesicularized ATP7B.