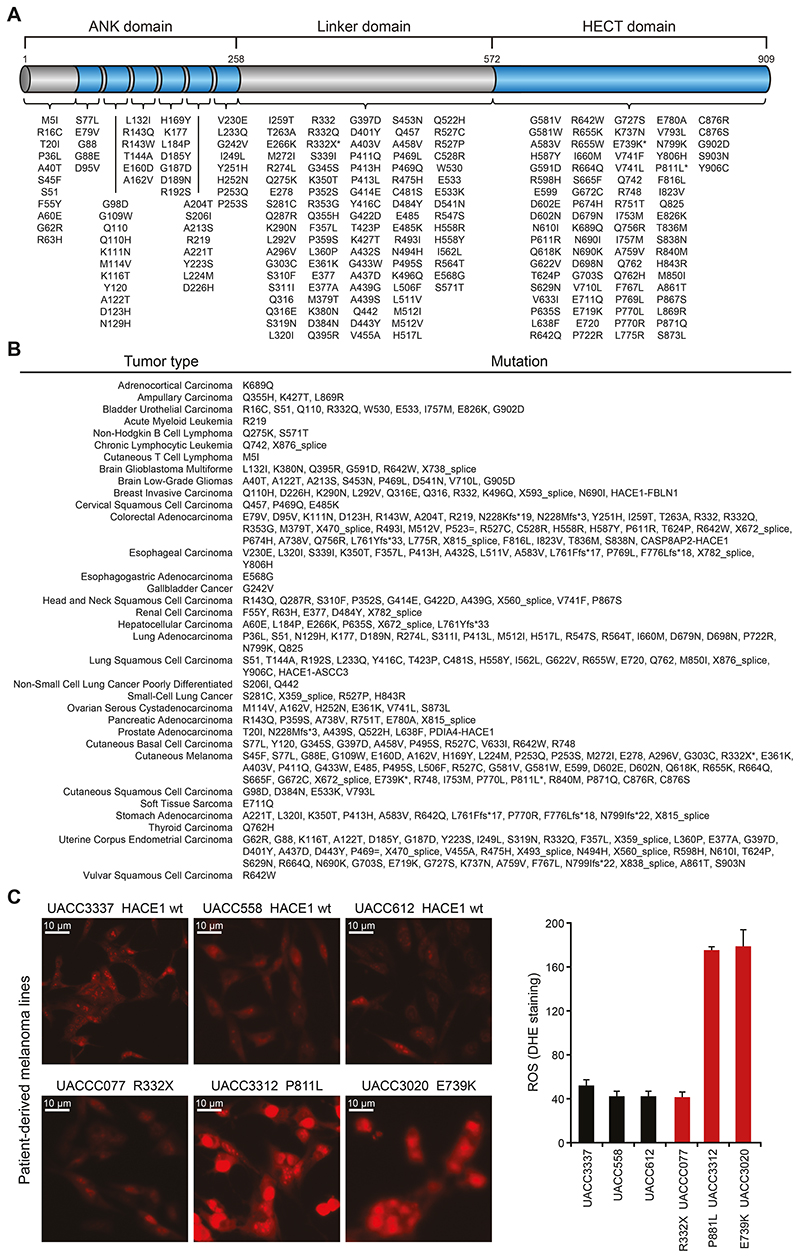
Figure 1. The HACE1 mutation landscape of human cancer.
(A) Intra-protein location of HACE1 mutations identified in human cancers as available from datasets on cBioPortal and TGen datasets. The asterisks highlight HACE1 mutations present in patient-derived melanoma cell lines used in (C). The different HACE1 domains are indicated. (B) HACE1 mutation profile in individual cancer types as available from the cBioPortal and TGen datasets. (C) Patient-derived human melanoma cell lines expressing endogenous wildtype HACE1 (wt) or mutated (R332X, P811L, E739K) HACE1, were analyzed for superoxide content by dihydroethidium (DHE) staining. Representative pictures of DHE-stained patient-derived melanoma cells (left) and the quantitative analysis of DHE fluorescence intensity in 200 cells/cell line (right) (Student’s two-tailed t-test, n=3) are shown. Red staining indicates the presence of reactive oxygen species (ROS). Scale bars, 20μm. Data in (C) are presented as mean values SEM.