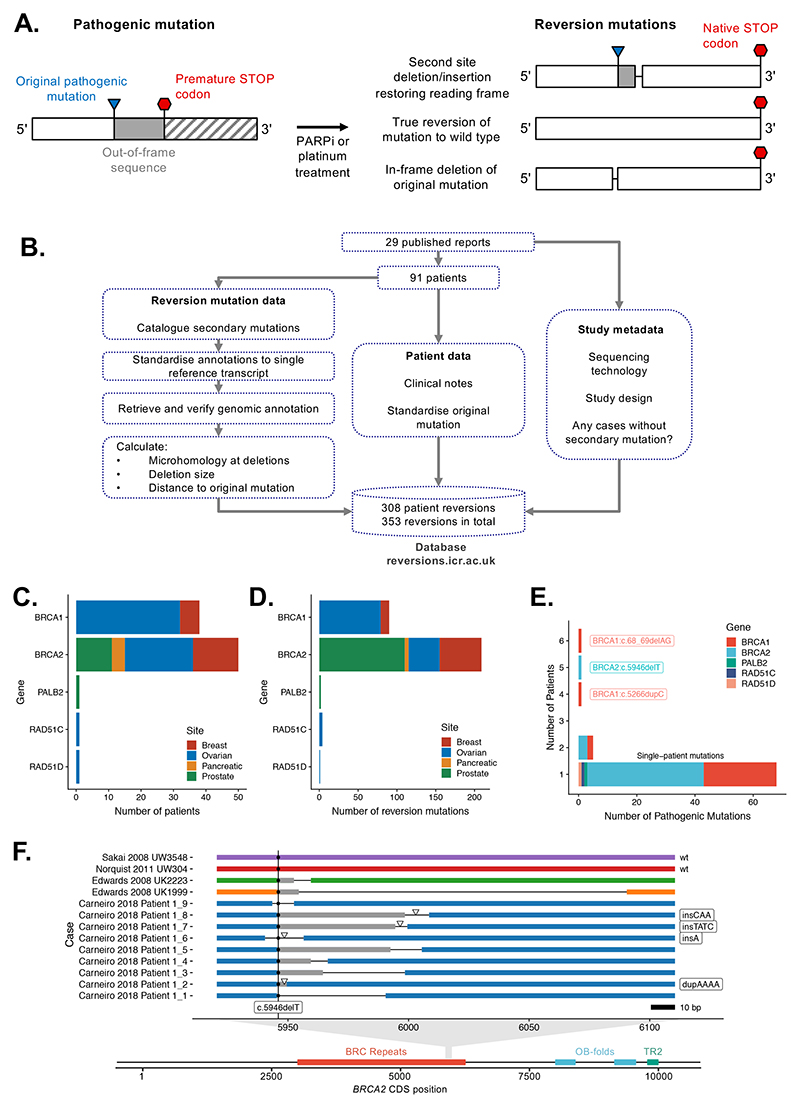
Figure 1. Collation, annotation and standardisation of HR gene reversion mutations.
A. Common architectures of HR gene reversion mutations associated with platinum or PARPi resistance. B. Workflow schematic illustrating the collation, annotation and standardisation of HR gene reversion mutations. C. Bar chart illustrating the primary tumor site in 91 patients with HR gene reversions described in the dataset. Patients are stratified by HR gene and by primary tumor site (see color key). D. Bar chart illustrating 308 reversion mutations in the dataset, stratified by HR gene and by primary tumor site. E. Bar chart illustrating that the majority of reversion mutations in the dataset arise from patients with different pathogenic mutations. Most patients (77%) had unique pathogenic mutations (annotated as “single-patient” mutations). Reversion cases from multiple patients with common Ashkenazi founder mutations, such as BRCA2:c.6174delT (c.5946delT in standardised nomenclature) and BRCA1:c.185delAG (c.68_69delAG), were also identified. F. Example of unique reversion events observed for multiple patients with a common founder mutation, BRCA2:c.6174delT (c.5496delT), represented on the BRCA2 coding sequence (CDS). Two true reversions to wild-type DNA sequence were observed in two different patients. Second site reversion mutations in other patients are also shown, colored by patient. Deletions are indicated by thin black lines. Sites of insertions are shown by triangles, with the inserted bases listed to the right. Out-of-frame sequence between pathogenic and reversion mutation is shaded in grey. The position of the pathogenic c.5946delT mutation is indicated by a vertical line.